Plant Science Research Weekly: January 22, 2021
Review: Molecular mechanisms involved in functional macroevolution of plant transcription factors
 Transcription factors (TFs) are very important actors through which evolution can operate. In every organism and system studied, starting with the seminal work of Jacob and Monod, they’ve been shown to be potent regulatory proteins. Here, Romani and Moreno review the contributions of plant transcription factors in plant evolution. Genomic studies have revealed that key innovations have been accompanied by TF family expansion and neofunctionalization. Here, the authors review functional studies that investigate TFs and gene-regulatory networks in Marchantia, Physcomitrium, and other newly-emerging non-angiosperm models. They conclude that in spite of millions of years of divergence, the functions of most TFs are largely conserved across plants, particularly those involved in developmental processes, but less so for those involved in environmental responses that may have been acquired largely independently. (Summary by Mary Williams @PlantTeaching) New Phytol. 10.1111/nph.17161
Transcription factors (TFs) are very important actors through which evolution can operate. In every organism and system studied, starting with the seminal work of Jacob and Monod, they’ve been shown to be potent regulatory proteins. Here, Romani and Moreno review the contributions of plant transcription factors in plant evolution. Genomic studies have revealed that key innovations have been accompanied by TF family expansion and neofunctionalization. Here, the authors review functional studies that investigate TFs and gene-regulatory networks in Marchantia, Physcomitrium, and other newly-emerging non-angiosperm models. They conclude that in spite of millions of years of divergence, the functions of most TFs are largely conserved across plants, particularly those involved in developmental processes, but less so for those involved in environmental responses that may have been acquired largely independently. (Summary by Mary Williams @PlantTeaching) New Phytol. 10.1111/nph.17161
Review: The genetic control of succulent leaf development
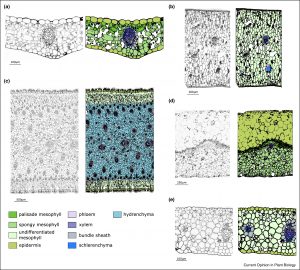 Succulence provides plants with the ability to store water and is therefore commonly associated with plants from arid environments such as the familiar Aloe and Agave. Here, Heyduk reviews the genetic control of leaf succulence. Succulence usually involves large, highly vacuolated cells, but not surprisingly, succulence takes many forms (e.g., whether or not all cells are enlarged, number of cell layers, patterns of vascular tissues). In this review, the author discusses the contributions to succulence of molecular players known from model plants that are involved in patterning, cell division and expansion. As she points out, the ability to introduce one or two cell layers capable of storing water could be a big advantage to rainfed crops that periodically experience water limitation. (Summary by Mary Williams @PlantTeaching). Curr. Opin. Plant Biol. 10.1016/j.pbi.2020.11.003
Succulence provides plants with the ability to store water and is therefore commonly associated with plants from arid environments such as the familiar Aloe and Agave. Here, Heyduk reviews the genetic control of leaf succulence. Succulence usually involves large, highly vacuolated cells, but not surprisingly, succulence takes many forms (e.g., whether or not all cells are enlarged, number of cell layers, patterns of vascular tissues). In this review, the author discusses the contributions to succulence of molecular players known from model plants that are involved in patterning, cell division and expansion. As she points out, the ability to introduce one or two cell layers capable of storing water could be a big advantage to rainfed crops that periodically experience water limitation. (Summary by Mary Williams @PlantTeaching). Curr. Opin. Plant Biol. 10.1016/j.pbi.2020.11.003
The Arabidopsis NOT4A E3 ligase promotes PGR3 expression and regulates chloroplast translation
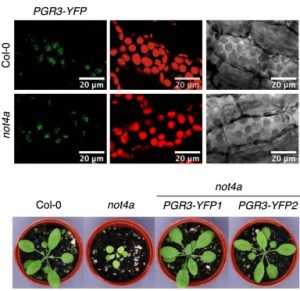 Pentatricopeptide repeat (PPR) domain containing proteins are nuclear encoded with functions in the chloroplast, but regulation of PPR gene expression in the nucleus before import to the chloroplast has not been well studied. Bailey et al. identified and characterized the ubiquitin ligase NOT4A and its regulatory function on PGR3, a PPR protein important for several photosynthetic processes. A not4a mutant displayed pale yellow coloration, delayed flowering, and starch depletion compared to wild-type. RNA sequencing and proteomics confirmed that several photosystem II and cytochrome b6f complex proteins are mis-regulated in the not4a mutant. Chloroplast 30 S ribosome subunit proteins were depleted in not4a although they were elevated in the RNA dataset. The not4a mutant was hypersensitive to the plastid ribosome inhibitor lincomycin, and wild-type plants treated with lincomycin phenocopied untreated not4a, suggesting that chloroplast mRNA translation is impaired in not4a mutant. Lastly, PGR3 expression was significantly downregulated in not4a, and PGR3 expression complements impaired chloroplast functions not4a mutant. Taken together, the research shows that NOT4A is essential for proper photosynthetic function and positively regulates PGR3 and chloroplast translation. (Summary by Toluwase Olukayode @toluxylic) Nature Comms. 10.1038/s41467-020-20506-4
Pentatricopeptide repeat (PPR) domain containing proteins are nuclear encoded with functions in the chloroplast, but regulation of PPR gene expression in the nucleus before import to the chloroplast has not been well studied. Bailey et al. identified and characterized the ubiquitin ligase NOT4A and its regulatory function on PGR3, a PPR protein important for several photosynthetic processes. A not4a mutant displayed pale yellow coloration, delayed flowering, and starch depletion compared to wild-type. RNA sequencing and proteomics confirmed that several photosystem II and cytochrome b6f complex proteins are mis-regulated in the not4a mutant. Chloroplast 30 S ribosome subunit proteins were depleted in not4a although they were elevated in the RNA dataset. The not4a mutant was hypersensitive to the plastid ribosome inhibitor lincomycin, and wild-type plants treated with lincomycin phenocopied untreated not4a, suggesting that chloroplast mRNA translation is impaired in not4a mutant. Lastly, PGR3 expression was significantly downregulated in not4a, and PGR3 expression complements impaired chloroplast functions not4a mutant. Taken together, the research shows that NOT4A is essential for proper photosynthetic function and positively regulates PGR3 and chloroplast translation. (Summary by Toluwase Olukayode @toluxylic) Nature Comms. 10.1038/s41467-020-20506-4
Cold translation: Ribosome-mediated translation inhibition senses cold stress in plants
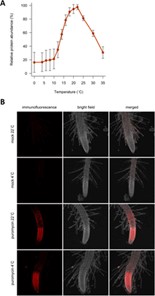 A drop in ambient temperature adversely affects plant growth and development. While the primary molecular pathway in response to cold stress involves the expression of CBF transcription factors, Guillaume-Schöpfer and colleagues found cold stress-induced inhibition of ribosome translation machinery to control CBF expression. To start, the rate of protein synthesis was found to be proportional to the decrease in temperature. The authors showed that cycloheximide (CHX) specifically induced rapid expression of CBF and other early cold-responsive genes. Strikingly, this CHX-mediated induction of cold-responsive genes required functional CALMODULIN-BINDING TRANSCRIPTION ACTIVATOR (CAMTA) proteins, which are the upstream regulators of CBFs. Further, the authors showed CHX-mediated increase in cytosolic and nuclear free calcium levels are required for CBF expression that may possibly work through the CAMTAs. Together, they suggest ribosomes to be “cryosensors” that control the early phase of gene expression in response to cold stress. (Summary by Pavithran Narayanan @pavi_narayanan). bioRxiv 10.1101/2020.12.07.414789v1.full
A drop in ambient temperature adversely affects plant growth and development. While the primary molecular pathway in response to cold stress involves the expression of CBF transcription factors, Guillaume-Schöpfer and colleagues found cold stress-induced inhibition of ribosome translation machinery to control CBF expression. To start, the rate of protein synthesis was found to be proportional to the decrease in temperature. The authors showed that cycloheximide (CHX) specifically induced rapid expression of CBF and other early cold-responsive genes. Strikingly, this CHX-mediated induction of cold-responsive genes required functional CALMODULIN-BINDING TRANSCRIPTION ACTIVATOR (CAMTA) proteins, which are the upstream regulators of CBFs. Further, the authors showed CHX-mediated increase in cytosolic and nuclear free calcium levels are required for CBF expression that may possibly work through the CAMTAs. Together, they suggest ribosomes to be “cryosensors” that control the early phase of gene expression in response to cold stress. (Summary by Pavithran Narayanan @pavi_narayanan). bioRxiv 10.1101/2020.12.07.414789v1.full
BONZAIs emerge as nodal regulators of osmotic stress responses
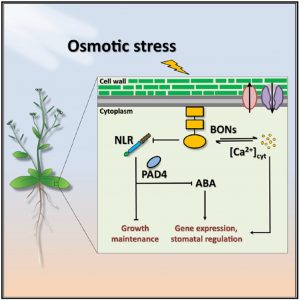 Plants launch their response to osmotic stress with a sudden spike in cytosolic Ca2+ levels, which subsequently leads to the accumulation of the phytohormone abscisic acid (ABA). This further initiates a chain of events including closure of stomata, large-scale transcriptional changes, retardation of overall growth, and the onset of early senescence. However, the mechanism by which the initial Ca2+ spike induced by osmotic stress leads to ABA accumulation remains poorly understood. In their recent study, Chen et al. identified the calcium-binding protein BONZAI1 (BON1) as a key molecular link between these events. Mutating BON1 alone or in combination with its functionally overlapping paralogs BON2 and BON3 not only hampered Ca2+ signaling but also reduced ABA accumulation, leading to severely compromised osmotic stress responses in Arabidopsis seedlings. The authors showed that BONs mediated the transcriptional changes of several adaptation genes in response to osmotic stress, a large proportion of them being regulated independently of ABA. Additionally, BONs played a crucial role in maintaining vegetative growth under osmotic stress, which also occurred largely in an ABA-independent manner. Therefore, BONs acted in the upstream of both ABA-dependent and ABA-independent responses, coordinating their activation under osmotic stress. Interestingly, the authors also found that BONs promoted osmotic stress responses by suppressing NLR receptor-mediated immune signaling, indicating their role in balancing the antagonism between abiotic and biotic stress responses to optimize the survival of plants. (Summary by Yadukrishnan Premachandran @yadukrishprem) Curr. Biol. 10.1016/j.cub.2020.09.016
Plants launch their response to osmotic stress with a sudden spike in cytosolic Ca2+ levels, which subsequently leads to the accumulation of the phytohormone abscisic acid (ABA). This further initiates a chain of events including closure of stomata, large-scale transcriptional changes, retardation of overall growth, and the onset of early senescence. However, the mechanism by which the initial Ca2+ spike induced by osmotic stress leads to ABA accumulation remains poorly understood. In their recent study, Chen et al. identified the calcium-binding protein BONZAI1 (BON1) as a key molecular link between these events. Mutating BON1 alone or in combination with its functionally overlapping paralogs BON2 and BON3 not only hampered Ca2+ signaling but also reduced ABA accumulation, leading to severely compromised osmotic stress responses in Arabidopsis seedlings. The authors showed that BONs mediated the transcriptional changes of several adaptation genes in response to osmotic stress, a large proportion of them being regulated independently of ABA. Additionally, BONs played a crucial role in maintaining vegetative growth under osmotic stress, which also occurred largely in an ABA-independent manner. Therefore, BONs acted in the upstream of both ABA-dependent and ABA-independent responses, coordinating their activation under osmotic stress. Interestingly, the authors also found that BONs promoted osmotic stress responses by suppressing NLR receptor-mediated immune signaling, indicating their role in balancing the antagonism between abiotic and biotic stress responses to optimize the survival of plants. (Summary by Yadukrishnan Premachandran @yadukrishprem) Curr. Biol. 10.1016/j.cub.2020.09.016
Ethylene response factors 15 and 16 trigger jasmonate biosynthesis in tomato during herbivore resistance
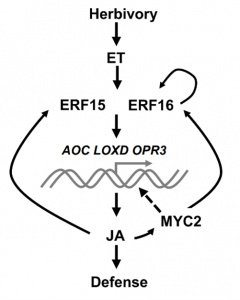 Crop damage and yield losses caused by herbivores have become major threats to global food security. Upon wounding and herbivory, plants rapidly accumulate high levels of jasmonates (JA). However, the mechanism underlying how JA biosynthesis is triggered by herbivore attack remains unclear. It is therefore essential to identify key regulatory components involved in herbivory-induced JA signaling and study their role in resistance. Hu et al. showed that tomato defense against the cotton bollworm Helicoverpa armigera (H. armigera) were orchestrated by both JA and ethylene. The authors identified two ethylene-induced ETHYLENE RESPONSE FACTOR (ERF) genes namely, ERF15 and ERF16, as transcription factors able to bind to the promoters of JA biosynthesis genes and trigger downstream JA signaling. Interestingly, plants lacking ERF15 and ERF16 displayed increased susceptibility to H. armigera due to reduced JA levels and JA biosynthesis gene expression. Interestingly, the JA-activated MYC2 and ERF16 were also able to bind the promoter of ERF16 to activate its expression. Altogether, Hu et al. identified a new layer of tomato defense regulation against H. armigera where ethylene-responsive ERF15 and ERF16 act as positive regulators of the JA burst in tomato in response to H. armigera. (Summary by Sarah Courbier @Scourbier) Plant Physiol. 10.1093/plphys/kiaa089
Crop damage and yield losses caused by herbivores have become major threats to global food security. Upon wounding and herbivory, plants rapidly accumulate high levels of jasmonates (JA). However, the mechanism underlying how JA biosynthesis is triggered by herbivore attack remains unclear. It is therefore essential to identify key regulatory components involved in herbivory-induced JA signaling and study their role in resistance. Hu et al. showed that tomato defense against the cotton bollworm Helicoverpa armigera (H. armigera) were orchestrated by both JA and ethylene. The authors identified two ethylene-induced ETHYLENE RESPONSE FACTOR (ERF) genes namely, ERF15 and ERF16, as transcription factors able to bind to the promoters of JA biosynthesis genes and trigger downstream JA signaling. Interestingly, plants lacking ERF15 and ERF16 displayed increased susceptibility to H. armigera due to reduced JA levels and JA biosynthesis gene expression. Interestingly, the JA-activated MYC2 and ERF16 were also able to bind the promoter of ERF16 to activate its expression. Altogether, Hu et al. identified a new layer of tomato defense regulation against H. armigera where ethylene-responsive ERF15 and ERF16 act as positive regulators of the JA burst in tomato in response to H. armigera. (Summary by Sarah Courbier @Scourbier) Plant Physiol. 10.1093/plphys/kiaa089
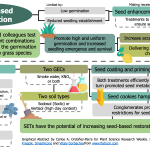
Seed enhancement technologies to improve germination and emergence of Australian native Poaceae
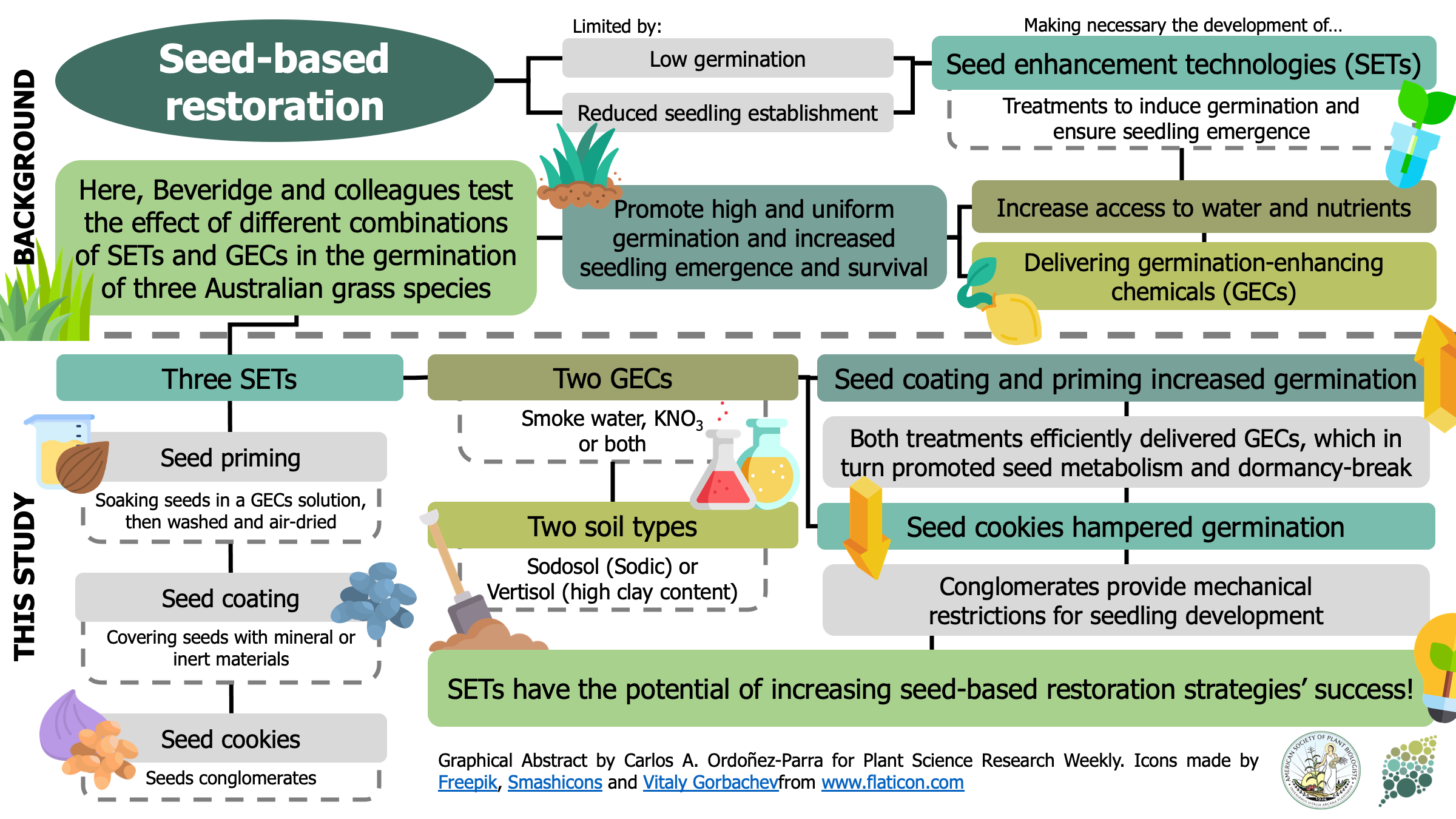 One of the most significant limitations in seed-based restoration projects is that some native species exhibit low recruitment in the field. As a result, various seed enhancement technologies (SETs) are being developed to improve germination and seedling emergence by facilitating access to nutrients and water. Moreover, they can include germination-enhancing chemicals (GECs) that help seeds overcome dormancy. In this research, Beveridge and colleagues test the effect of different combinations of SETs and GECs in the germination of three Australian grass species. Seeds were treated with three SETs (seed priming, seed coating, and seed cookies) in combination with two GECs (smoke water, KNO3, or both) and planted in pots with two contrasting soil types. Regardless of the soil, seed cookies reduced germination, probably because they provided mechanical restrictions to seedling development. In contrast, coating and priming seeds with at least one GEC significantly enhanced germination by presumably aiding seeds to break dormancy and resume metabolic activities. Therefore, this research shows the potential of SETs to increase the success of seed-based restoration strategies. Moreover, it provides a useful baseline for future efforts to widen the use of these technologies in other native species. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Seed Sci. Res. 10.1017/S0960258520000276
One of the most significant limitations in seed-based restoration projects is that some native species exhibit low recruitment in the field. As a result, various seed enhancement technologies (SETs) are being developed to improve germination and seedling emergence by facilitating access to nutrients and water. Moreover, they can include germination-enhancing chemicals (GECs) that help seeds overcome dormancy. In this research, Beveridge and colleagues test the effect of different combinations of SETs and GECs in the germination of three Australian grass species. Seeds were treated with three SETs (seed priming, seed coating, and seed cookies) in combination with two GECs (smoke water, KNO3, or both) and planted in pots with two contrasting soil types. Regardless of the soil, seed cookies reduced germination, probably because they provided mechanical restrictions to seedling development. In contrast, coating and priming seeds with at least one GEC significantly enhanced germination by presumably aiding seeds to break dormancy and resume metabolic activities. Therefore, this research shows the potential of SETs to increase the success of seed-based restoration strategies. Moreover, it provides a useful baseline for future efforts to widen the use of these technologies in other native species. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Seed Sci. Res. 10.1017/S0960258520000276