Plant Science Research Weekly: January 19, 2024
Review. Milestones in understanding phosphorus uptake, transport, sensing, use, and signaling
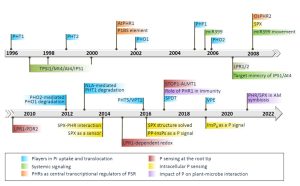 Phosphorus (P) is an essential nutrient and critical component of nucleic acids, phospholipids, and other molecules. Yang et al. provide a historical (since 1996) overview of the processes controlling its uptake and use. Plants take up P from the rhizosphere primarily in the form of orthophosphate (Pi). However, Pi in the soil often complexes with metal ions, limiting its availability. The discovery in 1996 of root-specific PHT1 phosphate transporters, localized in the plasma membrane and induced during Pi deficiency, initiated extensive exploration into this vital nutrient element. Subsequent identification of Pi transporters with expression in various tissues (e.g. stele, reproductive tissue, embryo) or subcellular organelles (e.g. chloroplasts/plastids, mitochondria, Golgi apparatus, vacuole) highlighted their role in mediating Pi uptake, cell-to-cell movement, and long-distance transport, thereby ensuring a balance between internal homeostasis and external supply and ultimately maximizing plant performance. Achieving this homeostasis demands a complex interplay of diverse and sophisticated molecular regulatory mechanisms including microRNAs, transcription factors, untranslated open reading frames, the ubiquitin ligase system, protein phosphorylation, and ligand-regulated protein-protein interactions. Furthermore, recent breakthroughs elucidating the structural-functional relationship of the regulatory PP-InsP/SPX/PHR complex not only enhance our understanding of the P signaling cascade but also unveil exciting opportunities for the interplay between P signaling and other crucial physiological processes, including nitrogen and iron homeostasis, and the dynamics of plant-microbe interactions. (Summary by Ching Chan @ntnuchanlab) Plant Cell 10.1093/plcell/koad326
Phosphorus (P) is an essential nutrient and critical component of nucleic acids, phospholipids, and other molecules. Yang et al. provide a historical (since 1996) overview of the processes controlling its uptake and use. Plants take up P from the rhizosphere primarily in the form of orthophosphate (Pi). However, Pi in the soil often complexes with metal ions, limiting its availability. The discovery in 1996 of root-specific PHT1 phosphate transporters, localized in the plasma membrane and induced during Pi deficiency, initiated extensive exploration into this vital nutrient element. Subsequent identification of Pi transporters with expression in various tissues (e.g. stele, reproductive tissue, embryo) or subcellular organelles (e.g. chloroplasts/plastids, mitochondria, Golgi apparatus, vacuole) highlighted their role in mediating Pi uptake, cell-to-cell movement, and long-distance transport, thereby ensuring a balance between internal homeostasis and external supply and ultimately maximizing plant performance. Achieving this homeostasis demands a complex interplay of diverse and sophisticated molecular regulatory mechanisms including microRNAs, transcription factors, untranslated open reading frames, the ubiquitin ligase system, protein phosphorylation, and ligand-regulated protein-protein interactions. Furthermore, recent breakthroughs elucidating the structural-functional relationship of the regulatory PP-InsP/SPX/PHR complex not only enhance our understanding of the P signaling cascade but also unveil exciting opportunities for the interplay between P signaling and other crucial physiological processes, including nitrogen and iron homeostasis, and the dynamics of plant-microbe interactions. (Summary by Ching Chan @ntnuchanlab) Plant Cell 10.1093/plcell/koad326
Review: CEP hormones at the nexus of nutrient acquisition and allocation, root development, and plant–microbe interactions
 C-TERMINALLY ENCODED PEPTIDE (CEP) is a multigene family of peptide hormones originally described as systemic long-distance signals in response to nitrogen limitation. Over the years since the discovery of these peptide hormones, the literature on CEP biology has expanded the repertoire of developmental and physiological roles in planta beyond nitrogen demand signaling. This review by Taleski et al. is a timely update on the multifaceted functions of CEP hormones in various biological processes including nutrient uptake, root development, and plant-microbe symbioses in angiosperms. Furthermore, the review features emerging themes and evidence on topics like plant immunity, where an atypical CEP member functions as novel immune-responsive phytocytokine to modulate plant health. The authors also discuss the specificity of different members of the CEP family. As an example, CEP2, but not other CEPs, seems to have a role in interactions with arbuscular mycorrhizae. CEP2 is downregulated by the presence of the fungi, promoting root growth. As CEP signaling is postulated to be conserved throughout all seed plants, it will be interesting to see if there is a common conserved signaling network across families and lineages. For example, CEP function in gymnosperms has not been investigated yet. This review provides a valuable update on CEP hormone function. (Summary by Marvin Jin @MarvinJYS) J. Exp. Bot. 10.1093/jxb/erad444
C-TERMINALLY ENCODED PEPTIDE (CEP) is a multigene family of peptide hormones originally described as systemic long-distance signals in response to nitrogen limitation. Over the years since the discovery of these peptide hormones, the literature on CEP biology has expanded the repertoire of developmental and physiological roles in planta beyond nitrogen demand signaling. This review by Taleski et al. is a timely update on the multifaceted functions of CEP hormones in various biological processes including nutrient uptake, root development, and plant-microbe symbioses in angiosperms. Furthermore, the review features emerging themes and evidence on topics like plant immunity, where an atypical CEP member functions as novel immune-responsive phytocytokine to modulate plant health. The authors also discuss the specificity of different members of the CEP family. As an example, CEP2, but not other CEPs, seems to have a role in interactions with arbuscular mycorrhizae. CEP2 is downregulated by the presence of the fungi, promoting root growth. As CEP signaling is postulated to be conserved throughout all seed plants, it will be interesting to see if there is a common conserved signaling network across families and lineages. For example, CEP function in gymnosperms has not been investigated yet. This review provides a valuable update on CEP hormone function. (Summary by Marvin Jin @MarvinJYS) J. Exp. Bot. 10.1093/jxb/erad444
Review: Stem cells for crop improvement
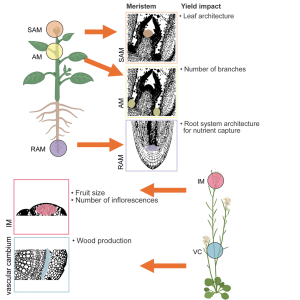 Plants, like animals, have small populations of stem cells capable of differentiating into other tissues, but in plants these stem cell populations are even more long-lived and versatile. Stem cells in plants include the meristems (shoot and root apical meristems, inflorescence and axillary meristems) as well as the vascular cambium. Collectively, they produce all the post-embryonic tissues of the plant. The biology of plant stem cells and how they can be manipulated for crop improvement is the topic of a review by Lindsay et al. Much of the review focuses on our current understanding of stem cell function and regulation, largely determined through genetic studies. The review also discusses applications of stem cell biology. Specifically, manipulation of stem cell actions can contribute to novel, beneficial plant architectures, such as increased inflorescence branching to produce more seeds, or increased ear size to produce more kernels. Additionally, manipulation of stem cell activities has been extremely helpful in accelerating the regeneration of plants from tissue culture following gene editing or transformation. (Summary by Mary Williams @PlantTeaching) Mol. Plant 10.1016/j.molp.2023.12.014
Plants, like animals, have small populations of stem cells capable of differentiating into other tissues, but in plants these stem cell populations are even more long-lived and versatile. Stem cells in plants include the meristems (shoot and root apical meristems, inflorescence and axillary meristems) as well as the vascular cambium. Collectively, they produce all the post-embryonic tissues of the plant. The biology of plant stem cells and how they can be manipulated for crop improvement is the topic of a review by Lindsay et al. Much of the review focuses on our current understanding of stem cell function and regulation, largely determined through genetic studies. The review also discusses applications of stem cell biology. Specifically, manipulation of stem cell actions can contribute to novel, beneficial plant architectures, such as increased inflorescence branching to produce more seeds, or increased ear size to produce more kernels. Additionally, manipulation of stem cell activities has been extremely helpful in accelerating the regeneration of plants from tissue culture following gene editing or transformation. (Summary by Mary Williams @PlantTeaching) Mol. Plant 10.1016/j.molp.2023.12.014
Review: Salicylic acid in plant immunity and beyond
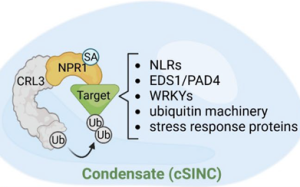 Salicylic acid (SA) is a pivotal natural compound in plant science and finds applications in herbal medicine; specifically, aspirin, the renowned anti-inflammatory drug and pain reliever, is a derivative of SA. As summarized in this review by Spoel and Dong, within plants SA serves as a crucial phytohormone essential for basal immunity and systemic acquired resistance (SAR). Its synthesis initiates in the chloroplast and increases in response to pathogen invasion. The activation of SA-signaling correlates positively with plant immunity, impacting both localized infections and distant tissues. A key molecular target for SA resides within the nonexpresser of PR proteins (NPRs), acting as authentic SA receptors. NPR3 and NPR4 play a pivotal role in spatially regulating NPR1, thereby mediating cell death to restrict infection. Consequently, SA exhibits a dual role: as a signal conferring protection and cell survival, and as a suicidal signal triggering cell death. The review also looks at the regulation of SAR. Although numerous biomolecules have been proposed to facilitate long-distance signaling, clear evidence via feeding experiments remains elusive. A recent preprint suggested that the extent of local reactive oxygen species induction might determine the intensity of SAR in distal tissues, shedding light on a three-decade-old query regarding how local infections lead to de novo SA synthesis in systemic tissues. Furthermore, the structural identification of NPR1 oligomerization, the unique property of NPR1 to form biomolecular condensates, and insights into CULLIN-dependent ubiquitin ligase complex assembly, have unveiled revolutionary insights into the molecular regulation of SA-dependent signaling in plant immunity. (Summary by Ching Chan @ntnuchanlab) Plant Cell 10.1093/plcell/koad329
Salicylic acid (SA) is a pivotal natural compound in plant science and finds applications in herbal medicine; specifically, aspirin, the renowned anti-inflammatory drug and pain reliever, is a derivative of SA. As summarized in this review by Spoel and Dong, within plants SA serves as a crucial phytohormone essential for basal immunity and systemic acquired resistance (SAR). Its synthesis initiates in the chloroplast and increases in response to pathogen invasion. The activation of SA-signaling correlates positively with plant immunity, impacting both localized infections and distant tissues. A key molecular target for SA resides within the nonexpresser of PR proteins (NPRs), acting as authentic SA receptors. NPR3 and NPR4 play a pivotal role in spatially regulating NPR1, thereby mediating cell death to restrict infection. Consequently, SA exhibits a dual role: as a signal conferring protection and cell survival, and as a suicidal signal triggering cell death. The review also looks at the regulation of SAR. Although numerous biomolecules have been proposed to facilitate long-distance signaling, clear evidence via feeding experiments remains elusive. A recent preprint suggested that the extent of local reactive oxygen species induction might determine the intensity of SAR in distal tissues, shedding light on a three-decade-old query regarding how local infections lead to de novo SA synthesis in systemic tissues. Furthermore, the structural identification of NPR1 oligomerization, the unique property of NPR1 to form biomolecular condensates, and insights into CULLIN-dependent ubiquitin ligase complex assembly, have unveiled revolutionary insights into the molecular regulation of SA-dependent signaling in plant immunity. (Summary by Ching Chan @ntnuchanlab) Plant Cell 10.1093/plcell/koad329
Comment: Unlocking the potential of agricultural biotechnology in Africa
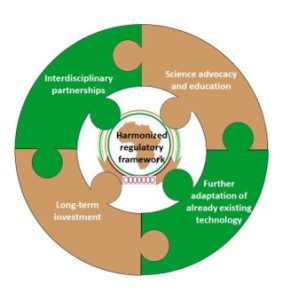 Agricultural Biotechnology has the potential to significantly increase production, alleviate hunger, and improve global food security. However, its adoption in Africa lags behind other regions, facing challenges rooted in strict and varying policies, inadequate infrastructure, and insufficient economic incentives for private-sector investment. Despite some successful initiatives in the past, the key to success lies in achieving a more extensive and widespread implementation. This paper identifies five practical opportunities for delivering and adopting agricultural biotechnology, suggesting specific actions to address the current challenges. A crucial aspect is the need for greater collaboration and the establishment of a unified market and regulatory environment. Incentives for incoming funding and grants from the private sector should be established, with a focus on long-term development, including talent training, retention, and infrastructure development and maintenance. Emphasizing ongoing education and communication between scientists and the public is vital for the successful adaptation and acceptance of new technologies. In light of these considerations, any new adaptation plan should prioritize education, transparency, and collaboration. This approach ensures the successful integration of agricultural biotechnology in Africa, ultimately leading to enhanced productivity and improved food security. (Summary by Villő Bernád) Nature Plants 10.1038/s41477-023-01604-9
Agricultural Biotechnology has the potential to significantly increase production, alleviate hunger, and improve global food security. However, its adoption in Africa lags behind other regions, facing challenges rooted in strict and varying policies, inadequate infrastructure, and insufficient economic incentives for private-sector investment. Despite some successful initiatives in the past, the key to success lies in achieving a more extensive and widespread implementation. This paper identifies five practical opportunities for delivering and adopting agricultural biotechnology, suggesting specific actions to address the current challenges. A crucial aspect is the need for greater collaboration and the establishment of a unified market and regulatory environment. Incentives for incoming funding and grants from the private sector should be established, with a focus on long-term development, including talent training, retention, and infrastructure development and maintenance. Emphasizing ongoing education and communication between scientists and the public is vital for the successful adaptation and acceptance of new technologies. In light of these considerations, any new adaptation plan should prioritize education, transparency, and collaboration. This approach ensures the successful integration of agricultural biotechnology in Africa, ultimately leading to enhanced productivity and improved food security. (Summary by Villő Bernád) Nature Plants 10.1038/s41477-023-01604-9
A novel starch granule size distribution in Arabidopsis thaliana is associated with differences in phosphorylation
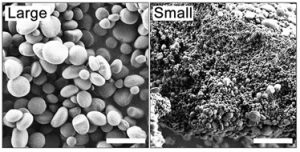 During the day, starch is synthesized in the chloroplasts of leaves and stored as starch granules, before being degraded in the subsequent night. In Arabidopsis thaliana, starch granules are discoid in shape. Some mutants have round or elongated granules, yet all these mutants have a homogenous starch granule morphology. Here Li et al. generated Arabidopsis thaliana double mutants for two genes involved in starch degradation – STARCH EXCESS 4 (SEX4) and DISPROPORTIONATING ENZYME2 (DPE2). Surprisingly there was a non-homogenous distribution of starch granules in the chloroplasts of dpe2sex4 mutants, with populations of large and small starch granules with 5-fold difference in size. Phosphorylation has a large effect on starch granules, thus the authors measured phosphate levels in the large and small granules. When accounting for surface area, small granules had a 37% reduction in phosphate content compared to large granules. There were also differences in phosphate distribution, with phosphate distributed throughout the small granules compared to being mainly on the surface of the large granules. Hence, the authors have discovered a novel starch granule size distribution in Arabidopsis thaliana dep2sex4 mutants which may be attributed to differences in starch phosphorylation. (Summary by Rose McNelly @Rose_McN) Plant Physiol. 10.1093/plphys/kiad656
During the day, starch is synthesized in the chloroplasts of leaves and stored as starch granules, before being degraded in the subsequent night. In Arabidopsis thaliana, starch granules are discoid in shape. Some mutants have round or elongated granules, yet all these mutants have a homogenous starch granule morphology. Here Li et al. generated Arabidopsis thaliana double mutants for two genes involved in starch degradation – STARCH EXCESS 4 (SEX4) and DISPROPORTIONATING ENZYME2 (DPE2). Surprisingly there was a non-homogenous distribution of starch granules in the chloroplasts of dpe2sex4 mutants, with populations of large and small starch granules with 5-fold difference in size. Phosphorylation has a large effect on starch granules, thus the authors measured phosphate levels in the large and small granules. When accounting for surface area, small granules had a 37% reduction in phosphate content compared to large granules. There were also differences in phosphate distribution, with phosphate distributed throughout the small granules compared to being mainly on the surface of the large granules. Hence, the authors have discovered a novel starch granule size distribution in Arabidopsis thaliana dep2sex4 mutants which may be attributed to differences in starch phosphorylation. (Summary by Rose McNelly @Rose_McN) Plant Physiol. 10.1093/plphys/kiad656
Shooting yourself in the foot: Obligate biotrophic pathogen induces protective microbiome in Arabidopsis
 Obligate plant pathogens cannot survive without their host plant. Therefore, understanding how the plant and its microbiome respond to the obligate biotrophs has real challenges in making sure that the response that is being monitored is not an artifact of the experimental step up. A recent study by Goossens et al. showed how the phyllosphere bacterial microbiome of Arabidopsis thaliana plants is affected by the infection of the obligate biotrophic downy mildew Hyaloperonospora arabidopsidis (Hpa). Upon inoculation with Hpa, Arabidopsis plants studied in the Netherlands and Germany developed very similar microbiota, which the authors named Hpa-associated microbiota (HAM). Surprisingly, the researchers showed that the HAM were not helping the pathogen, but instead reduced the disease symptoms of the plants by suppressing Hpa sporulation. When plants were sown in pathogen-conditioned soil, HAM bacteria accumulated in the phyllsophere, showing that plant rhizosphere can recruit these beneficial strains and maintain them on their phyllosphere. It remains to be shown how exactly the Arabidopsis plants can select for this beneficial microbiome. (Summary by Maria Constantin @meconstantin001). Nature Microbiology 10.1038/s41564-023-01502-y
Obligate plant pathogens cannot survive without their host plant. Therefore, understanding how the plant and its microbiome respond to the obligate biotrophs has real challenges in making sure that the response that is being monitored is not an artifact of the experimental step up. A recent study by Goossens et al. showed how the phyllosphere bacterial microbiome of Arabidopsis thaliana plants is affected by the infection of the obligate biotrophic downy mildew Hyaloperonospora arabidopsidis (Hpa). Upon inoculation with Hpa, Arabidopsis plants studied in the Netherlands and Germany developed very similar microbiota, which the authors named Hpa-associated microbiota (HAM). Surprisingly, the researchers showed that the HAM were not helping the pathogen, but instead reduced the disease symptoms of the plants by suppressing Hpa sporulation. When plants were sown in pathogen-conditioned soil, HAM bacteria accumulated in the phyllsophere, showing that plant rhizosphere can recruit these beneficial strains and maintain them on their phyllosphere. It remains to be shown how exactly the Arabidopsis plants can select for this beneficial microbiome. (Summary by Maria Constantin @meconstantin001). Nature Microbiology 10.1038/s41564-023-01502-y
PSKR1 balances the plant growth–defence trade-off in the rhizosphere microbiome
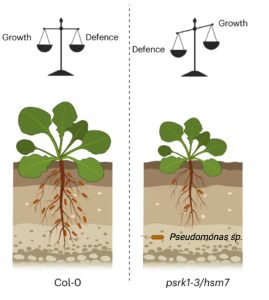 Plants are colonized by numerous beneficial microorganisms in the rhizosphere, including Pseudomonas fluorescens, that provide benefits including nutrient acquisition and pathogen protection. Host plants must tune their immune systems to restrict microbial overgrowth, while avoiding overstimulation of the immune system (autoimmunity). However, the genetic regulator that integrates rhizosphere microbial cues and autoimmune response is largely unknown. Using mutant screening, bulk segregant analyses, overexpression lines, and whole genome sequencing for autoimmune Arabidopsis mutants, Song et al. discovered that PHYTOSULFOKINE RECEPTOR 1 (PSKR1) mutants display autoimmunity, characterized by stunted growth and overactive immune responses in response to P.fluorecense. To determine the crosstalk between PSKR1 and the rhizosphere microbiome, the researchers conducted 16S microbiome profiling followed by a synthetic community-based (29 members SynCom) experiment, both of which indicated a largely specific effect of PSKR1 towards Pseudomonas and vice-versa. The authors then performed transcriptional profiling (RNA-seq) of Col-0 and psrk1-3 and found that PSKR1 suppressed salicylic acid (SA) mediated defense responses. Additional studies showed that Pseudomonas, and not other bacteria, secrete small molecule(s) that might induce PSRK1 (thereby suppressing SA-mediated defense responses) to promote colonization and balance growth-defense tradeoffs. This study demonstrates the importance of PSRK1 in controlling defense responses and promoting interactions with beneficial Pseudomonas and thus offers strategies to engineer the rhizosphere microbiome to sustainably enhance plant health. (Summary by Arijit Mukherjee @ArijitM61745830) Nature Plants 10.1038/s41477-023-01539-1.
Plants are colonized by numerous beneficial microorganisms in the rhizosphere, including Pseudomonas fluorescens, that provide benefits including nutrient acquisition and pathogen protection. Host plants must tune their immune systems to restrict microbial overgrowth, while avoiding overstimulation of the immune system (autoimmunity). However, the genetic regulator that integrates rhizosphere microbial cues and autoimmune response is largely unknown. Using mutant screening, bulk segregant analyses, overexpression lines, and whole genome sequencing for autoimmune Arabidopsis mutants, Song et al. discovered that PHYTOSULFOKINE RECEPTOR 1 (PSKR1) mutants display autoimmunity, characterized by stunted growth and overactive immune responses in response to P.fluorecense. To determine the crosstalk between PSKR1 and the rhizosphere microbiome, the researchers conducted 16S microbiome profiling followed by a synthetic community-based (29 members SynCom) experiment, both of which indicated a largely specific effect of PSKR1 towards Pseudomonas and vice-versa. The authors then performed transcriptional profiling (RNA-seq) of Col-0 and psrk1-3 and found that PSKR1 suppressed salicylic acid (SA) mediated defense responses. Additional studies showed that Pseudomonas, and not other bacteria, secrete small molecule(s) that might induce PSRK1 (thereby suppressing SA-mediated defense responses) to promote colonization and balance growth-defense tradeoffs. This study demonstrates the importance of PSRK1 in controlling defense responses and promoting interactions with beneficial Pseudomonas and thus offers strategies to engineer the rhizosphere microbiome to sustainably enhance plant health. (Summary by Arijit Mukherjee @ArijitM61745830) Nature Plants 10.1038/s41477-023-01539-1.
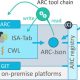
PLANTdataHUB- One plant research data platform to rule them all
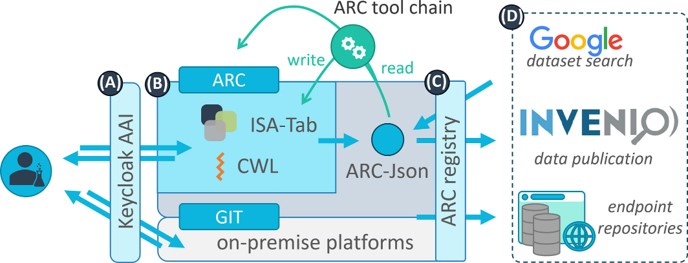 Research Data Management for plant scientists just got easier with the newly released PLANTdataHub. In this platform, you can register your data which will be organized in Annotated Research Contexts (ARCs). ARCS are Findable Accessible Interoperable Reproducible (FAIR) digital Objects which are the best practices for storing plant data. PLANTdataHub allows you to version your data, collaborate, reuse, reference, and publish your data all from one place. This platform will help shape the future of Plant Research Data Management. Have a look at this free platform and consider using it as an individual or a lab here. (Summary by Maria Constantin @meconstantin001). Plant J. 10.1111/tpj.16474
Research Data Management for plant scientists just got easier with the newly released PLANTdataHub. In this platform, you can register your data which will be organized in Annotated Research Contexts (ARCs). ARCS are Findable Accessible Interoperable Reproducible (FAIR) digital Objects which are the best practices for storing plant data. PLANTdataHub allows you to version your data, collaborate, reuse, reference, and publish your data all from one place. This platform will help shape the future of Plant Research Data Management. Have a look at this free platform and consider using it as an individual or a lab here. (Summary by Maria Constantin @meconstantin001). Plant J. 10.1111/tpj.16474