Plant Science Research Weekly: January 13, 2023
An RNA world
 Annual Reviews editions typically start with a brief autobiographical essay by a noted scientist. The 2023 edition of Annual Reviews in Plant Biology includes a profile of David Baulcombe, the well-known scientist whose work was foundational in revealing the roles of small RNAs in gene regulation and viral suppression. Baulcombe started his PhD in 1973, and his narrative reads like a history of the ideas and methods that shaped the past 50 years of plant molecular biology, particularly the growing recognition of its RNA world. Throughout the article, Baulcombe champions both basic and applied plant research, discussing the need for policies that support “sustainable intensification” in agriculture (appreciating its nuances), and seeking greater recognition that plants are excellent models for basic biology (suggesting that this message “should be shouted for the rooftops”). His article concludes with a closer look at the dance between host and pathogens, as well as the contributions of transposons in plant evolution. Throughout, he reveals his deep understanding of the important questions of our discipline, recognizes the contributions of his mentors and proteges to these findings, and demonstrates why he is widely perceived as one of the true stars of plant science. (Summary by Mary Williams @PlantTeaching) Annu. Rev. Plant Biol. 10.1146/annurev-arplant-070622-021021
Annual Reviews editions typically start with a brief autobiographical essay by a noted scientist. The 2023 edition of Annual Reviews in Plant Biology includes a profile of David Baulcombe, the well-known scientist whose work was foundational in revealing the roles of small RNAs in gene regulation and viral suppression. Baulcombe started his PhD in 1973, and his narrative reads like a history of the ideas and methods that shaped the past 50 years of plant molecular biology, particularly the growing recognition of its RNA world. Throughout the article, Baulcombe champions both basic and applied plant research, discussing the need for policies that support “sustainable intensification” in agriculture (appreciating its nuances), and seeking greater recognition that plants are excellent models for basic biology (suggesting that this message “should be shouted for the rooftops”). His article concludes with a closer look at the dance between host and pathogens, as well as the contributions of transposons in plant evolution. Throughout, he reveals his deep understanding of the important questions of our discipline, recognizes the contributions of his mentors and proteges to these findings, and demonstrates why he is widely perceived as one of the true stars of plant science. (Summary by Mary Williams @PlantTeaching) Annu. Rev. Plant Biol. 10.1146/annurev-arplant-070622-021021
Plant Cell Focus Issue on climate change and plant abiotic stress biology
 The January 2023 issue of The Plant Cell includes a focus issue on “Climate change and plant stress: From genes to ecosystems”. This timely focus issue has in it commentaries, reviews, and research articles on the topic. I particularly enjoyed reading the two reviews that include short perspectives from multiple authors. The first addresses how plant science can help to mitigate climate change and enhance food security, spanning from carbon sequestration in soils to improving photosynthesis and developing climate resilient crops. The second highlights key open questions in plant abiotic stress biology, from questions about how plants sense and integrate stress signals to the contributions of root plasticity in climate resilience. Several other articles look at how current breeding tools are being deployed in the study of stress tolerance and signaling, as well as the exploitation of this knowledge towards the production of climate-resilient crops. Most of the articles in this Focus Issue are available to read without a subscription. (Summary by Mary Williams @PlantTeaching) Plant Cell https://academic.oup.com/plcell/issue/35/1.
The January 2023 issue of The Plant Cell includes a focus issue on “Climate change and plant stress: From genes to ecosystems”. This timely focus issue has in it commentaries, reviews, and research articles on the topic. I particularly enjoyed reading the two reviews that include short perspectives from multiple authors. The first addresses how plant science can help to mitigate climate change and enhance food security, spanning from carbon sequestration in soils to improving photosynthesis and developing climate resilient crops. The second highlights key open questions in plant abiotic stress biology, from questions about how plants sense and integrate stress signals to the contributions of root plasticity in climate resilience. Several other articles look at how current breeding tools are being deployed in the study of stress tolerance and signaling, as well as the exploitation of this knowledge towards the production of climate-resilient crops. Most of the articles in this Focus Issue are available to read without a subscription. (Summary by Mary Williams @PlantTeaching) Plant Cell https://academic.oup.com/plcell/issue/35/1.
Perspective: Multiple mechanism behind plant bending
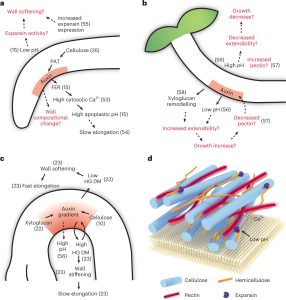 Bending or folding in plants is influenced by different factors such as environmental conditions (nutrient, water, light, gravity), abiotic or biotic stress, cell wall properties, and cell differential growth. Jonsson et al. explain how bending is achieved by considering molecular mechanisms, mathematical models, and biophysical aspects. The plant bending process can be classified into ‘fountain-like’ and ‘hinge-like’ bending, with the difference being the cell dynamic of the bent region over time; in hinge-like bending, the same cells always occupy the site of bending, whereas in fountain-like bending the curved region is displaced in the organ over time. The apical hook (which forms when the hypocotyl bends) is an example of ‘fountain-like’ bending, with bending enabled by differential cell growth on the outer and inner sides. The phytohormone auxin has been suggested to play an essential role in the initial bending of the apical hook, together with cell wall components. Moreover, ethylene, brassinosteroid, and gibberellin signaling also can be involved in the tissue bending process. In addition to biochemical signals, mechanical signals such as turgor pressure and mechanical conflicts between cells and tissues complement the folding complexity in plant cells. Mathematical models can help us to understand cellular mechanisms regarding biochemical and mechanical signals. This review provides an overview of bending mechanisms at different levels of regulation along with perspectives to understand the folding process. (Summary by Andrea Gómez-Felipe @andreagomezfe) Nature Plants 10.1038/s41477-022-01310-y
Bending or folding in plants is influenced by different factors such as environmental conditions (nutrient, water, light, gravity), abiotic or biotic stress, cell wall properties, and cell differential growth. Jonsson et al. explain how bending is achieved by considering molecular mechanisms, mathematical models, and biophysical aspects. The plant bending process can be classified into ‘fountain-like’ and ‘hinge-like’ bending, with the difference being the cell dynamic of the bent region over time; in hinge-like bending, the same cells always occupy the site of bending, whereas in fountain-like bending the curved region is displaced in the organ over time. The apical hook (which forms when the hypocotyl bends) is an example of ‘fountain-like’ bending, with bending enabled by differential cell growth on the outer and inner sides. The phytohormone auxin has been suggested to play an essential role in the initial bending of the apical hook, together with cell wall components. Moreover, ethylene, brassinosteroid, and gibberellin signaling also can be involved in the tissue bending process. In addition to biochemical signals, mechanical signals such as turgor pressure and mechanical conflicts between cells and tissues complement the folding complexity in plant cells. Mathematical models can help us to understand cellular mechanisms regarding biochemical and mechanical signals. This review provides an overview of bending mechanisms at different levels of regulation along with perspectives to understand the folding process. (Summary by Andrea Gómez-Felipe @andreagomezfe) Nature Plants 10.1038/s41477-022-01310-y
Review: The establishment of plants following long-distance dispersal
 Seed dispersal is a critical ecological process with far-reaching implications. On rare occasions, seeds can be dispersed over vast distances, far beyond the range of the species. These events, known as long-distance dispersal, have profound evolutionary implications by expanding species distribution and triggering speciation. However, a comprehensive framework for understanding is still missing. In this exciting review, Wu and colleagues provide a first-of-its-kind synthesis of long-distance dispersal and its implication for species biogeography and evolution. The authors identify six factors that shape this process: 1) propagule pressure, 2) functional traits, 3) extreme events and anthropogenic disturbances, 4) predators, competitors, and mutualists, 5) niche flexibility and 6) Allee effects. Notably, the authors emphasize the role of humans in long-distance dispersal, as human activities allow the movement of seeds all over the globe. As a result, studying long-distance dispersal should be a priority and a promising research area in the Anthropocene. The authors also outline simple formulas to estimate the probability of colonizing new sites. As a result, the review provides a theoretical and quantitative foundation for future advancements in this research area. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Trends Ecol. Evol. 10.1016/j.tree.2022.11.003
Seed dispersal is a critical ecological process with far-reaching implications. On rare occasions, seeds can be dispersed over vast distances, far beyond the range of the species. These events, known as long-distance dispersal, have profound evolutionary implications by expanding species distribution and triggering speciation. However, a comprehensive framework for understanding is still missing. In this exciting review, Wu and colleagues provide a first-of-its-kind synthesis of long-distance dispersal and its implication for species biogeography and evolution. The authors identify six factors that shape this process: 1) propagule pressure, 2) functional traits, 3) extreme events and anthropogenic disturbances, 4) predators, competitors, and mutualists, 5) niche flexibility and 6) Allee effects. Notably, the authors emphasize the role of humans in long-distance dispersal, as human activities allow the movement of seeds all over the globe. As a result, studying long-distance dispersal should be a priority and a promising research area in the Anthropocene. The authors also outline simple formulas to estimate the probability of colonizing new sites. As a result, the review provides a theoretical and quantitative foundation for future advancements in this research area. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Trends Ecol. Evol. 10.1016/j.tree.2022.11.003
The CIPK23 protein kinase represses SLAC1-type anion channels in Arabidopsis guard cells and stimulates stomatal opening
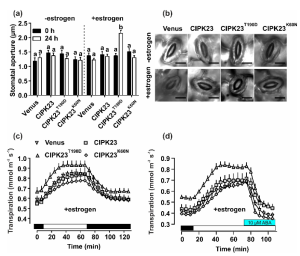 Guard cells are responsible for the opening and closing of stomata through changes in osmotic content and turgor pressure. These changes occur in response to activation or inactivation of ion transport proteins that are in turn regulated by protein kinase and phosphatase networks. The kinase network comprises different families, such as CBL-Interacting Protein Kinases (CIPKs). CIPK23 is thought to open stomata through the stimulation of K+ uptake channels in guard cells. However, some studies show that CIPK23 activates S-type anion channels SLAC1 and SLAH3, causing stomatal closure. In this work, Huang et al., addressed these contrasting data by generating different estrogen-inducible Arabidopsis lines for guard cell specific expression of different versions of CIPK23. In particular, they used CIPK23 together with the phosphorylation mimic T190D. The authors showed that the expression of CIPK23T190D in guard cells stimulates stomatal opening through the inactivation of SLAC1-type anion channels. This work sheds light into how CIPK23 control several transport proteins and therefore stomatal movements. (Summary by Eva Maria Gomez Alvarez, @eva_ga96) New Phytologist. 10.1111/nph.18708
Guard cells are responsible for the opening and closing of stomata through changes in osmotic content and turgor pressure. These changes occur in response to activation or inactivation of ion transport proteins that are in turn regulated by protein kinase and phosphatase networks. The kinase network comprises different families, such as CBL-Interacting Protein Kinases (CIPKs). CIPK23 is thought to open stomata through the stimulation of K+ uptake channels in guard cells. However, some studies show that CIPK23 activates S-type anion channels SLAC1 and SLAH3, causing stomatal closure. In this work, Huang et al., addressed these contrasting data by generating different estrogen-inducible Arabidopsis lines for guard cell specific expression of different versions of CIPK23. In particular, they used CIPK23 together with the phosphorylation mimic T190D. The authors showed that the expression of CIPK23T190D in guard cells stimulates stomatal opening through the inactivation of SLAC1-type anion channels. This work sheds light into how CIPK23 control several transport proteins and therefore stomatal movements. (Summary by Eva Maria Gomez Alvarez, @eva_ga96) New Phytologist. 10.1111/nph.18708
Contribution of strigolactones to maize kernel domesticated phenotype
 We’ve all seen striking photographs comparing the modern maize (Zea mays spp. mays L.) ear to the seed head of its wild-grass ancestor teosinte (Zea mays spp. parviglumis). Besides the huge increase in size, one of the features that allows us to munch on “corn on the cob” is the absence of the hard, shell-like coverings on each kernel. Previous work identified Teosinte glume architecture 1 (Tga1), a gene that has a role in this domestication and encodes a SPL-type transcription factor. Maize carries a dominant gain-of-function mutation in Tga1 that has pleiotropic effects but does not alone account for the differences in maize and teosinte ear structures. Here, Guan et al. describe the additional involvement of strigolactone hormones in maize ear formation. They looked at strigolactone synthesis mutants (ccd8), in which several facets of maize development including ear formation are affected; in many ways, the SL-deficient maize ears, particularly when combined with the ancestral tga1 allele, strongly resemble those of teosinte. As the authors report, these SL-mediated effects occur both through interactions with TGA1 (via protein sequestration) and independently. They conclude that “SLs have central roles in releasing kernels from restrictive maternal encasement and coordinating other factors that increase kernel size, physical support, and their exposure on the grain-bearing surface.” (Summary by Mary Williams @PlantTeaching) Plant Cell 10.1093/plcell/koac370
We’ve all seen striking photographs comparing the modern maize (Zea mays spp. mays L.) ear to the seed head of its wild-grass ancestor teosinte (Zea mays spp. parviglumis). Besides the huge increase in size, one of the features that allows us to munch on “corn on the cob” is the absence of the hard, shell-like coverings on each kernel. Previous work identified Teosinte glume architecture 1 (Tga1), a gene that has a role in this domestication and encodes a SPL-type transcription factor. Maize carries a dominant gain-of-function mutation in Tga1 that has pleiotropic effects but does not alone account for the differences in maize and teosinte ear structures. Here, Guan et al. describe the additional involvement of strigolactone hormones in maize ear formation. They looked at strigolactone synthesis mutants (ccd8), in which several facets of maize development including ear formation are affected; in many ways, the SL-deficient maize ears, particularly when combined with the ancestral tga1 allele, strongly resemble those of teosinte. As the authors report, these SL-mediated effects occur both through interactions with TGA1 (via protein sequestration) and independently. They conclude that “SLs have central roles in releasing kernels from restrictive maternal encasement and coordinating other factors that increase kernel size, physical support, and their exposure on the grain-bearing surface.” (Summary by Mary Williams @PlantTeaching) Plant Cell 10.1093/plcell/koac370
eINTACT – An effective system for isolating effector-recipient cells in plant tissues
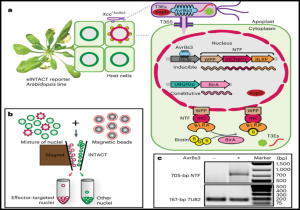 Bacteria can cause plant diseases by secreting small molecules called effectors. Studies of these effectors on host plants mostly have not accounted for the cellular complexity of plant tissues. The difficulty of detecting and isolating effector-recipient cells means that scientists mostly use bulk-infected host tissues or transgenic expression of effector genes to study the functions of bacterial effectors. These methods become limited in cases where the effector functions are dose-dependent, cell-specific, and/or change based on spatiotemporal distribution. In this study, You et al. introduce a technique that allows the purification of nuclei from Arabidopsis plant cells that have received Xanthomonas bacteria effectors. This technique, called eINTACT (effector-inducible isolation of nuclei tagged in specific cell types), uses a bacterial effector to activate transcription of a nuclear envelope-targeting protein (NTF), which tags effector-recipient nuclei for easy purification. Yuan et al. used eINTACT to study the effect of a Xanthomonas effector protein (XopD) on the dynamics of host gene expression and hormone signalling. The eINTACT system provides a unique opportunity to improve our understanding of the in planta functions of effectors from numerous gram-negative plant bacteria in native infection contexts. (Summary by Abdulkabir Omeiza Abdulmalik @Omeiza_PlantDoc) Nature Plants 10.1038/s41477-022-01302-y
Bacteria can cause plant diseases by secreting small molecules called effectors. Studies of these effectors on host plants mostly have not accounted for the cellular complexity of plant tissues. The difficulty of detecting and isolating effector-recipient cells means that scientists mostly use bulk-infected host tissues or transgenic expression of effector genes to study the functions of bacterial effectors. These methods become limited in cases where the effector functions are dose-dependent, cell-specific, and/or change based on spatiotemporal distribution. In this study, You et al. introduce a technique that allows the purification of nuclei from Arabidopsis plant cells that have received Xanthomonas bacteria effectors. This technique, called eINTACT (effector-inducible isolation of nuclei tagged in specific cell types), uses a bacterial effector to activate transcription of a nuclear envelope-targeting protein (NTF), which tags effector-recipient nuclei for easy purification. Yuan et al. used eINTACT to study the effect of a Xanthomonas effector protein (XopD) on the dynamics of host gene expression and hormone signalling. The eINTACT system provides a unique opportunity to improve our understanding of the in planta functions of effectors from numerous gram-negative plant bacteria in native infection contexts. (Summary by Abdulkabir Omeiza Abdulmalik @Omeiza_PlantDoc) Nature Plants 10.1038/s41477-022-01302-y
Targeted A-to-G base editing of chloroplast DNA in plants
 The chloroplast has its own genome which encodes proteins needed for chloroplast function. Editing these genes via CRISPR-Cas9 is difficult due to challenges in targeting guide RNAs to the chloroplast. Instead, base editors can be used, but these were originally developed for cytosine to thymine edits in mitochondria. Here, Mok et al. have used similar ideas to develop a chloroplast targeted adenine to guanine (A to G) base editor. Their editor consists of pairs of chloroplast targeted TALE (transcription activator-like effector) DNA-binding arrays linked to deaminases. When designed to target the photosynthetic gene psaA in Arabidopsis, there was specific editing of adenine to guanine with up to 99% efficiency, and this was accompanied by the expected albino phenotype. Interestingly, one plant was mosaic-like, with both green and pale leaves. The pale leaves had edits at the psaA start codon in 95% of cases, whereas in the green leaves the editing efficiency was 0.1%, demonstrating that the pale phenotype is in fact caused by the editing. Remarkably, these edits persisted in the subsequent generation and in some cases without the presence of the transgene. Therefore, the specific heritable edits generated by this base editor will make it easier to alter the chloroplast genome to study gene function. (Summary by Rose McNelly @Rose_McN) Nature Plants 10.1038/s41477-022-01279-8
The chloroplast has its own genome which encodes proteins needed for chloroplast function. Editing these genes via CRISPR-Cas9 is difficult due to challenges in targeting guide RNAs to the chloroplast. Instead, base editors can be used, but these were originally developed for cytosine to thymine edits in mitochondria. Here, Mok et al. have used similar ideas to develop a chloroplast targeted adenine to guanine (A to G) base editor. Their editor consists of pairs of chloroplast targeted TALE (transcription activator-like effector) DNA-binding arrays linked to deaminases. When designed to target the photosynthetic gene psaA in Arabidopsis, there was specific editing of adenine to guanine with up to 99% efficiency, and this was accompanied by the expected albino phenotype. Interestingly, one plant was mosaic-like, with both green and pale leaves. The pale leaves had edits at the psaA start codon in 95% of cases, whereas in the green leaves the editing efficiency was 0.1%, demonstrating that the pale phenotype is in fact caused by the editing. Remarkably, these edits persisted in the subsequent generation and in some cases without the presence of the transgene. Therefore, the specific heritable edits generated by this base editor will make it easier to alter the chloroplast genome to study gene function. (Summary by Rose McNelly @Rose_McN) Nature Plants 10.1038/s41477-022-01279-8
A successful genetic transformation procedure for turnip (Brassica rapa var. rapa)
 Genome editing has strong potential for crop improvement, but its application is restricted by low plant regeneration frequencies and genotype dependencies. Despite its agricultural significance, research on turnip (Brassica rapa var. rapa) is hindered by a low efficiency of transformation and shoot regeneration. In this study, Yuanyuan et al. introduced a procedure for stable genetic transformation and gene editing of turnip. They used the Agrobacterium-mediated expression of a development regulator gene encodling the WUSCHEL transcription factor (BrrWUSa) as an estrogen-inducible construct (pER8-BrrWUSa) to enhance fertile shoot regeneration in turnip. This transformation procedure was used to successfully carry out gene editing of BrrTCP4b gene in turnip. Taken together, this method may play an important role in the functional characterization of genes for turnip and provides a powerful tool for the future improvement of turnip by CRISPR/Cas-mediated gene editing. (Summary by Abdulkabir Omeiza Abdulmalik @Omeiza_PlantDoc) Plant Methods 10.1186/s13007-022-00931-w
Genome editing has strong potential for crop improvement, but its application is restricted by low plant regeneration frequencies and genotype dependencies. Despite its agricultural significance, research on turnip (Brassica rapa var. rapa) is hindered by a low efficiency of transformation and shoot regeneration. In this study, Yuanyuan et al. introduced a procedure for stable genetic transformation and gene editing of turnip. They used the Agrobacterium-mediated expression of a development regulator gene encodling the WUSCHEL transcription factor (BrrWUSa) as an estrogen-inducible construct (pER8-BrrWUSa) to enhance fertile shoot regeneration in turnip. This transformation procedure was used to successfully carry out gene editing of BrrTCP4b gene in turnip. Taken together, this method may play an important role in the functional characterization of genes for turnip and provides a powerful tool for the future improvement of turnip by CRISPR/Cas-mediated gene editing. (Summary by Abdulkabir Omeiza Abdulmalik @Omeiza_PlantDoc) Plant Methods 10.1186/s13007-022-00931-w
Flash flaming is a valid seed enhancement for a diverse range of species and seed morphologies
 One of the main challenges of seed-based restoration is incorporating seeds from as wide a variety of species as possible. However, the seeds of some species have shapes or appendages that make it challenging to use specialized machinery for direct seeding, which hinders large-scale restoration projects and may lead to the exclusion of certain species. In this research, Ling and colleagues show that flash flaming (i.e., sequentially exposing seeds to short-duration heating events) is a promising tool for processing seeds of 16 species widely used in ecological restoration in Western Australia. The technique reduced the volume and increased the flowability of all species’ seeds without negatively affecting most species’ germination or seed viability. In fact, the germination of two species increased after its flaming, suggesting it contributed to breaking their dormancy. The optimal flaming parameters –such as the number of flames, flame type, and duration– were species-specific, highlighting the need to standardize the technique with every new species. Still, this study provides exciting results that could spark and facilitate the use of more species in seed-based restoration efforts. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Seed Sci. Technol. 10.15258/sst.2022.50.3.10
One of the main challenges of seed-based restoration is incorporating seeds from as wide a variety of species as possible. However, the seeds of some species have shapes or appendages that make it challenging to use specialized machinery for direct seeding, which hinders large-scale restoration projects and may lead to the exclusion of certain species. In this research, Ling and colleagues show that flash flaming (i.e., sequentially exposing seeds to short-duration heating events) is a promising tool for processing seeds of 16 species widely used in ecological restoration in Western Australia. The technique reduced the volume and increased the flowability of all species’ seeds without negatively affecting most species’ germination or seed viability. In fact, the germination of two species increased after its flaming, suggesting it contributed to breaking their dormancy. The optimal flaming parameters –such as the number of flames, flame type, and duration– were species-specific, highlighting the need to standardize the technique with every new species. Still, this study provides exciting results that could spark and facilitate the use of more species in seed-based restoration efforts. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Seed Sci. Technol. 10.15258/sst.2022.50.3.10



