Plant Science Research Weekly: February 18th, 2022
Carbon flux through photosynthesis and central carbon metabolism show distinct patterns between algae, C3, and C4 plants
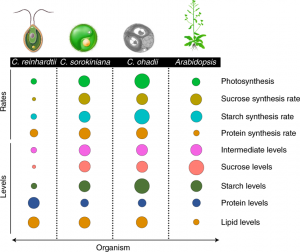 Photosynthesis is an attractive target for improving crop yields, and tailoring downstream photosynthesis-associated metabolism is a relatively unexplored path for achieving this. Chlorella ohadii is the fastest growing photosynthetic organism identified, has high photosynthetic rates, and can survive extreme illumination levels. Here, Treves et al. show how higher metabolic flux in C. ohadii could explain this fast growth compared to two other algae, Chlamydomonas reinhardtii and Chlorella sorokiniana, as well as differences in C.ohadii grown in low versus high light. They also demonstrate that all three algae display increased fluxes compared to the plants Arabidopsis (C3) and maize (C4). The authors used a microfluidics system for rapid pulse-chase 13CO2 labelling experiments and measured labelling kinetics over both short (5 – 40 s) and longer (15 – 300 min) time scales. The first measured 20 metabolic intermediates of the Calvin Benson Cycle, and the second measured flux into downstream sugars, intermediates of the tricarboxylic acid cycle and amino acids. K-means clustering on the enrichment kinetics in these experiments explored trends in metabolic fluxes grouped into four clusters for both experiments. Rates of protein and lipid synthesis were also highest in the Chlorella species. The higher anaplerotic flux (the flow of fixed carbon directed towards replacing TCA cycle intermediates) in algae compared to plants could support the increased carbon supply needed for the higher rate of protein synthesis and growth. (Summary by @Jiaaawen) Nature Plants 10.1038/s41477-021-01042-5
Photosynthesis is an attractive target for improving crop yields, and tailoring downstream photosynthesis-associated metabolism is a relatively unexplored path for achieving this. Chlorella ohadii is the fastest growing photosynthetic organism identified, has high photosynthetic rates, and can survive extreme illumination levels. Here, Treves et al. show how higher metabolic flux in C. ohadii could explain this fast growth compared to two other algae, Chlamydomonas reinhardtii and Chlorella sorokiniana, as well as differences in C.ohadii grown in low versus high light. They also demonstrate that all three algae display increased fluxes compared to the plants Arabidopsis (C3) and maize (C4). The authors used a microfluidics system for rapid pulse-chase 13CO2 labelling experiments and measured labelling kinetics over both short (5 – 40 s) and longer (15 – 300 min) time scales. The first measured 20 metabolic intermediates of the Calvin Benson Cycle, and the second measured flux into downstream sugars, intermediates of the tricarboxylic acid cycle and amino acids. K-means clustering on the enrichment kinetics in these experiments explored trends in metabolic fluxes grouped into four clusters for both experiments. Rates of protein and lipid synthesis were also highest in the Chlorella species. The higher anaplerotic flux (the flow of fixed carbon directed towards replacing TCA cycle intermediates) in algae compared to plants could support the increased carbon supply needed for the higher rate of protein synthesis and growth. (Summary by @Jiaaawen) Nature Plants 10.1038/s41477-021-01042-5
OsMADS14 and NF-YB1 cooperate in the direct activation of OsAGPL2 and Waxy during starch synthesis in rice endosperm
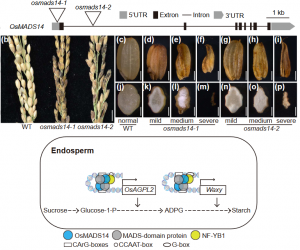 Rice grain is mainly composed of starch, which provides the necessary energy and sugars for successful germination and seedling development. Since rice is the most widely consumed crop, studying the regulators that control starch synthesis is crucial to ensure food security for more than half the world‘s population. In monocotyledonous species like rice, FUL-like proteins have been suggested to play a role during grain development, which includes several MADS-domain transcription factors (OsMADS). In this article, Feng et al. dissect the regulatory mechanisms for which OsMADS14 regulates the synthesis of starch during the grain filling process in rice. First, they found that two OsMADS14 loss-of-function mutant lines produce severely defective grain phenotypes. Through an RNA-Seq analysis, they identified genes repressed in the mutant lines including ADP-GLUCOSE PYROPHOSPHORYLASE LARGE SUBUNIT 2 (OsAGPL2) and Waxy (Wx). Both genes are directly regulated by OsMADS14. Through protein-protein interaction techniques NF-YB1 (a seed-specific transcription factor) was proposed to associate with OsMADS14 to directly promote OsAGPL2 and Wx gene expression. This work shows how the OsMADS14/ NF-YB1 complex directly regulates core starch synthesis genes. (Summary by Eva Maria Gomez Alvarez, @eva_ga96) New Phytol. 10.1111/nph.17990
Rice grain is mainly composed of starch, which provides the necessary energy and sugars for successful germination and seedling development. Since rice is the most widely consumed crop, studying the regulators that control starch synthesis is crucial to ensure food security for more than half the world‘s population. In monocotyledonous species like rice, FUL-like proteins have been suggested to play a role during grain development, which includes several MADS-domain transcription factors (OsMADS). In this article, Feng et al. dissect the regulatory mechanisms for which OsMADS14 regulates the synthesis of starch during the grain filling process in rice. First, they found that two OsMADS14 loss-of-function mutant lines produce severely defective grain phenotypes. Through an RNA-Seq analysis, they identified genes repressed in the mutant lines including ADP-GLUCOSE PYROPHOSPHORYLASE LARGE SUBUNIT 2 (OsAGPL2) and Waxy (Wx). Both genes are directly regulated by OsMADS14. Through protein-protein interaction techniques NF-YB1 (a seed-specific transcription factor) was proposed to associate with OsMADS14 to directly promote OsAGPL2 and Wx gene expression. This work shows how the OsMADS14/ NF-YB1 complex directly regulates core starch synthesis genes. (Summary by Eva Maria Gomez Alvarez, @eva_ga96) New Phytol. 10.1111/nph.17990
Shielding the oil reserves: the scutellum as a source of chemical defenses
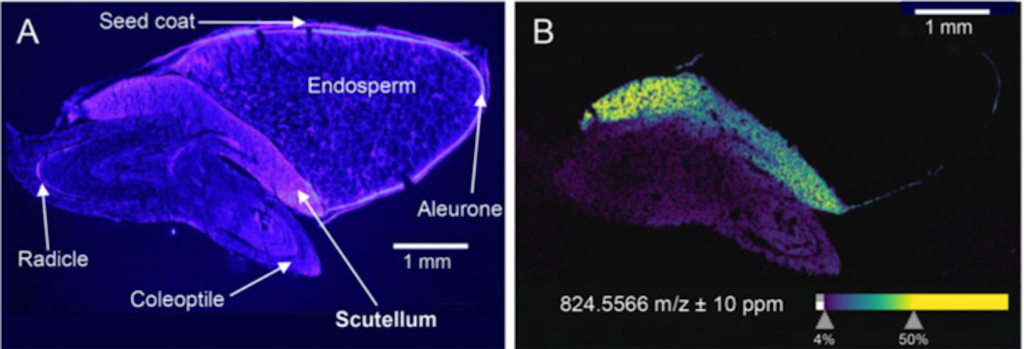 If you work with grasses, you are probably familiar with the scutellum –the shield-like cotyledon typical of seeds from these plants. This structure has a renowned role in transferring nutrients to the growing embryo. However, Murphy and colleagues show us that not only does this structure look like a shield, but it also protects the seedling from pathogens by producing and storing a myriad of defense compounds. Some examples of these substances include zealexins, benzoxazinoids and phenolamide derivates, which might be produced constitutively or in response to pathogen attacks. Notably, several of them are unique to the scutellum and even species-specific. Parallel metabolite and RNA-Seq analyses in maize seedlings show that these defense compounds are produced in the scutellum after germination. Therefore, these exciting findings pave the way for a fascinating new dimension of seedling biotic interactions. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Plant Physiol. 10.1093/plphys/kiac038
If you work with grasses, you are probably familiar with the scutellum –the shield-like cotyledon typical of seeds from these plants. This structure has a renowned role in transferring nutrients to the growing embryo. However, Murphy and colleagues show us that not only does this structure look like a shield, but it also protects the seedling from pathogens by producing and storing a myriad of defense compounds. Some examples of these substances include zealexins, benzoxazinoids and phenolamide derivates, which might be produced constitutively or in response to pathogen attacks. Notably, several of them are unique to the scutellum and even species-specific. Parallel metabolite and RNA-Seq analyses in maize seedlings show that these defense compounds are produced in the scutellum after germination. Therefore, these exciting findings pave the way for a fascinating new dimension of seedling biotic interactions. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Plant Physiol. 10.1093/plphys/kiac038
Structural organization of the spongy mesophyll
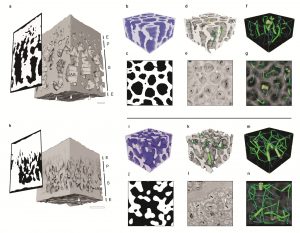 Despite a large variation in leaf morphology, laminar leaf anatomy is largely conserved, comprising two developmentally distinct mesophyll tissues, the palisade and spongy mesophyll. Spongy mesophyll cells along with intercellular spaces form an interconnected network to maximize the surface area for light capture and gas exchange. However, the structural organization of spongy mesophyll is relatively understudied due to the technical limitations of deep tissue imaging. Borsuk et al. analysed the 3D cellular geometry of spongy mesophyll in 40 species belonging to 30 genera using X-ray micro- computed tomography imaging. Contrary to the general perception of the spongy mesophyll as a random assembly of irregularly shaped cells, the authors observed two divergent phenotypes – the honeycomb and the non-honeycomb topology. The plants with honeycomb topology have larger cells, lower mesophyll surface area per unit volume (SAmes/Vmes) and lesser maximum photosynthetic rate (Amax). However, the honeycomb tessellation satisfies the multi-functional demands with economical construction costs, which suggests that two distinct topologies are optimized for either photosynthetic performance or minimal resource investment. The authors highlight flow rate directionality, cell arm length, cell packing density, and the characteristic minimum vein spacing as important structural drivers of the spongy mesophyll. This study shows that the structural organization of spongy mesophyll is a key trait influencing the leaf economy. (Summary by Prakshi Aneja @PrakshiAneja) New Phytol. 10.1111/nph.17971
Despite a large variation in leaf morphology, laminar leaf anatomy is largely conserved, comprising two developmentally distinct mesophyll tissues, the palisade and spongy mesophyll. Spongy mesophyll cells along with intercellular spaces form an interconnected network to maximize the surface area for light capture and gas exchange. However, the structural organization of spongy mesophyll is relatively understudied due to the technical limitations of deep tissue imaging. Borsuk et al. analysed the 3D cellular geometry of spongy mesophyll in 40 species belonging to 30 genera using X-ray micro- computed tomography imaging. Contrary to the general perception of the spongy mesophyll as a random assembly of irregularly shaped cells, the authors observed two divergent phenotypes – the honeycomb and the non-honeycomb topology. The plants with honeycomb topology have larger cells, lower mesophyll surface area per unit volume (SAmes/Vmes) and lesser maximum photosynthetic rate (Amax). However, the honeycomb tessellation satisfies the multi-functional demands with economical construction costs, which suggests that two distinct topologies are optimized for either photosynthetic performance or minimal resource investment. The authors highlight flow rate directionality, cell arm length, cell packing density, and the characteristic minimum vein spacing as important structural drivers of the spongy mesophyll. This study shows that the structural organization of spongy mesophyll is a key trait influencing the leaf economy. (Summary by Prakshi Aneja @PrakshiAneja) New Phytol. 10.1111/nph.17971
Microtubule-based perception of mechanical conflicts controls plant organ morphogenesis
 Morphogenesis is a critical process in plants and animals that allows a cell, tissue, or organism to reach its final shape. In Arabidopsis thaliana, the formation of lateral root primordia (LRP) is an excellent example of organ morphogenesis that involves cell remodeling throughout differential growth of an internal tissue. In this study, Stöckle et al. used live-cell imaging and cell type-specific genetic perturbations to show how differences in cortical microtubule organization between the inner and outer sides of the endodermis control LRP initiation. Before LRP initiation, cortical microtubules are more anisotropic on the side of the cell facing inwards and more disorganized on the other side. During LRP initiation, cortical microtubules on the inner side become disorganized. The reorganization of the cortical microtubules in the endodermal cells requires SHORT HYPOCOTYL2 (SHY2) and MICROTUBULE ASSOCIATED PROTEIN 70-5 (MAP70-5). map70-5 mutants display no particular organization of endodermal cortical microtubules and show flattened LRP. Stöckle et al. show that MAP70-5 is required for: i) the organization of cortical microtubules on the inner side of the endodermis, ii) remodeling of the endodermis during LRP initiation, and iii) LRP development and morphogenesis. The authors propose that cortical microtubules and MAP70-5 integrate the mechanical signals of LRP organogenesis. (Summary by Andrea Gómez-Felipe @andreagomezfe) Sci. Advanc. 10.1126/sciadv.abm4974
Morphogenesis is a critical process in plants and animals that allows a cell, tissue, or organism to reach its final shape. In Arabidopsis thaliana, the formation of lateral root primordia (LRP) is an excellent example of organ morphogenesis that involves cell remodeling throughout differential growth of an internal tissue. In this study, Stöckle et al. used live-cell imaging and cell type-specific genetic perturbations to show how differences in cortical microtubule organization between the inner and outer sides of the endodermis control LRP initiation. Before LRP initiation, cortical microtubules are more anisotropic on the side of the cell facing inwards and more disorganized on the other side. During LRP initiation, cortical microtubules on the inner side become disorganized. The reorganization of the cortical microtubules in the endodermal cells requires SHORT HYPOCOTYL2 (SHY2) and MICROTUBULE ASSOCIATED PROTEIN 70-5 (MAP70-5). map70-5 mutants display no particular organization of endodermal cortical microtubules and show flattened LRP. Stöckle et al. show that MAP70-5 is required for: i) the organization of cortical microtubules on the inner side of the endodermis, ii) remodeling of the endodermis during LRP initiation, and iii) LRP development and morphogenesis. The authors propose that cortical microtubules and MAP70-5 integrate the mechanical signals of LRP organogenesis. (Summary by Andrea Gómez-Felipe @andreagomezfe) Sci. Advanc. 10.1126/sciadv.abm4974
Cell-by-cell dissection of phloem development links a maturation gradient to cell specialization
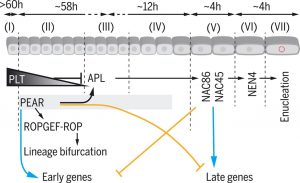 In this seminal paper, Roszak et al. present a deeper understanding of the development of the protophloem cell lineage. The differentiation process from the phloem stem cell to the enucleated cell state spans approximately 19 cells. Using live imaging, FACS sorting and single-cell RNA sequencing, the authors gain information about the transcriptomes of these cells and show that they can be grouped into seven distinct developmental stages. Interestingly, progression through these stages is associated with sharp transcriptomic changes realized through gene regulatory networks with mutual inhibition modules. The linage bifurcation of metaphloem and procambium cell files, taking place in stage II, depends on lineage-specific PHLOEM EARLY DNA-BINDING-WITH-ONE-FINGER (PEAR) transcription factors that activate distinct ROPGEF-ROP signaling to control asymmetric cell divisions. The transition from cell division to cell differentiation (stage II to IV) is dependent on a PLETHORA protein gradient directly repressing ALTERED PHLOEM DEVELOPMENT (APL) which functions in promoting cell maturation. In contrast, PEAR transcription factors promote phloem differentiation through activation of APL, initiating early differentiation. By functionally analyzing the interplay of key transcription factors, this paper provides a detailed understanding of developmental transitions and maturation processes in phloem development. (Summary by Aylin S Haas @AylinHaas_) Science 10.1126/science.aba5531
In this seminal paper, Roszak et al. present a deeper understanding of the development of the protophloem cell lineage. The differentiation process from the phloem stem cell to the enucleated cell state spans approximately 19 cells. Using live imaging, FACS sorting and single-cell RNA sequencing, the authors gain information about the transcriptomes of these cells and show that they can be grouped into seven distinct developmental stages. Interestingly, progression through these stages is associated with sharp transcriptomic changes realized through gene regulatory networks with mutual inhibition modules. The linage bifurcation of metaphloem and procambium cell files, taking place in stage II, depends on lineage-specific PHLOEM EARLY DNA-BINDING-WITH-ONE-FINGER (PEAR) transcription factors that activate distinct ROPGEF-ROP signaling to control asymmetric cell divisions. The transition from cell division to cell differentiation (stage II to IV) is dependent on a PLETHORA protein gradient directly repressing ALTERED PHLOEM DEVELOPMENT (APL) which functions in promoting cell maturation. In contrast, PEAR transcription factors promote phloem differentiation through activation of APL, initiating early differentiation. By functionally analyzing the interplay of key transcription factors, this paper provides a detailed understanding of developmental transitions and maturation processes in phloem development. (Summary by Aylin S Haas @AylinHaas_) Science 10.1126/science.aba5531
Shoot and root single cell sequencing reveals tissue- and daytime-specific transcriptome profiles
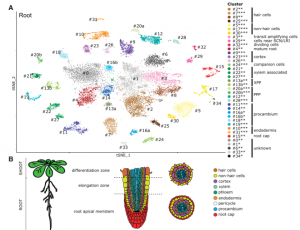 Many biological processes are controlled by the circadian clock in plants, yet studies often don’t report sampling timepoints, leaving much to be discovered about the plant circadian rhythm. Apelt et al. have utilised single cell RNA sequencing to examine almost 70,000 Arabidopsis cells from above and below ground tissues at day-end and night-end timepoints to reveal how the circadian clock influences the plant transcriptome. Transcriptomic changes in subterranean and above ground tissues varied with sampling period, leading to identification of day and night tissue-specific markers. MERCY1, a novel marker gene for dividing cells in both shoot and root tissues, may also influence flowering time, bolting and root development. This work provides the research community with a high-resolution transcriptomic map of single cells from shoot tissue for day and night and root tissue at an end of night timepoint. (Summary by Orla Sherwood @orlasherwood) Plant Physiol. 10.1093/plphys/kiab537
Many biological processes are controlled by the circadian clock in plants, yet studies often don’t report sampling timepoints, leaving much to be discovered about the plant circadian rhythm. Apelt et al. have utilised single cell RNA sequencing to examine almost 70,000 Arabidopsis cells from above and below ground tissues at day-end and night-end timepoints to reveal how the circadian clock influences the plant transcriptome. Transcriptomic changes in subterranean and above ground tissues varied with sampling period, leading to identification of day and night tissue-specific markers. MERCY1, a novel marker gene for dividing cells in both shoot and root tissues, may also influence flowering time, bolting and root development. This work provides the research community with a high-resolution transcriptomic map of single cells from shoot tissue for day and night and root tissue at an end of night timepoint. (Summary by Orla Sherwood @orlasherwood) Plant Physiol. 10.1093/plphys/kiab537
Ethylene augments root hypoxia tolerance through amelioration of reactive oxygen species and growth cessation
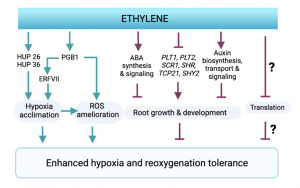 Flooding presents a dual threat to plants; not only is mitochondrial respiration supressed during the resulting hypoxic conditions, but once the floodwaters recede, the return to ambient oxygen levels triggers a cascade of harmful reaction oxygen species (ROS). One of the most well-studied responses to flooding stress is the accumulation of the gaseous hormone ethylene. While the core transcriptional machinery responsible for this response has been identified in Arabidopsis, the downstream pathways involved in hypoxia acclimation remain unclear. In this study, the authors show that pre-treating Arabidopsis plants with ethylene before exposing them to hypoxic and then re-oxygenated conditions reduces cell death in root tip cells. To reveal the mechanisms behind this, they conducted transcriptomic and proteomic analysis, showing that ethylene modulates mitochondrial metabolism, protein turnover, and ROS homeostasis during hypoxic conditions. This ethylene-induced ROS tolerance appears to be at least partially mediated by the proteins PGB1 and PRT6, which have previously been linked to flooding and general abiotic stress tolerance across multiple species. Ethylene also limits root growth, via its effect on auxin synthesis, and the authors show that this growth repression has a direct effect on hypoxia tolerance. (Summary by Rory Burke @rorby95) bioRxiv 10.1101/2022.01.21.477196
Flooding presents a dual threat to plants; not only is mitochondrial respiration supressed during the resulting hypoxic conditions, but once the floodwaters recede, the return to ambient oxygen levels triggers a cascade of harmful reaction oxygen species (ROS). One of the most well-studied responses to flooding stress is the accumulation of the gaseous hormone ethylene. While the core transcriptional machinery responsible for this response has been identified in Arabidopsis, the downstream pathways involved in hypoxia acclimation remain unclear. In this study, the authors show that pre-treating Arabidopsis plants with ethylene before exposing them to hypoxic and then re-oxygenated conditions reduces cell death in root tip cells. To reveal the mechanisms behind this, they conducted transcriptomic and proteomic analysis, showing that ethylene modulates mitochondrial metabolism, protein turnover, and ROS homeostasis during hypoxic conditions. This ethylene-induced ROS tolerance appears to be at least partially mediated by the proteins PGB1 and PRT6, which have previously been linked to flooding and general abiotic stress tolerance across multiple species. Ethylene also limits root growth, via its effect on auxin synthesis, and the authors show that this growth repression has a direct effect on hypoxia tolerance. (Summary by Rory Burke @rorby95) bioRxiv 10.1101/2022.01.21.477196
Convergent evolution of red nectar in vertebrate-pollinated flowers
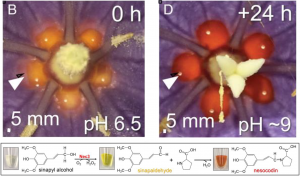 Nesocodon mauritianus is about one out of 70 plant species that produce colored nectar. The nectar of Nesocodon starts off yellow in newly opened flowers and progresses to blood-red in color as the flowers mature. The authors showed that the red colored nectar is both visible and attractive to geckos, the presumed pollinator of this and other Mauritian species. The authors identified sinapaldehyde in the nectar, which when conjugated to proline under alkalizing conditions results a red pigment named nesocodin. Nesocodin is synthesized from sinapyl alcohol to sinapaldehyde by an enzyme belonging to the glucose-methanol-choline oxidoreductase protein superfamily. A carbonic anhydrase increases the pH of the nectar to more alkaline conditions, allowing for the nonenzymatic conjugation of proline to sinapaldehyde to give nesocodin. Hydrogen peroxide is a by-product of sinapaldehyde formation that can break down the pigment; however, it is removed by a third enzyme, a catalase. The distantly related Jaltomata herrerae has red nectar and is pollinated by hummingbirds. The authors found nesocodin in the nectar of J. herrerae, as well as a carbonic anhydrase and an oxidoreductase for nesocodin biosynthesis. These findings inidicate that production of pigmented nectar in these two distantly-related vertebrate-pollinated flowers is a case of convergent evolution. (Summary by Kristen Van Gelder) PNAS 10.1073/pnas.2114420119
Nesocodon mauritianus is about one out of 70 plant species that produce colored nectar. The nectar of Nesocodon starts off yellow in newly opened flowers and progresses to blood-red in color as the flowers mature. The authors showed that the red colored nectar is both visible and attractive to geckos, the presumed pollinator of this and other Mauritian species. The authors identified sinapaldehyde in the nectar, which when conjugated to proline under alkalizing conditions results a red pigment named nesocodin. Nesocodin is synthesized from sinapyl alcohol to sinapaldehyde by an enzyme belonging to the glucose-methanol-choline oxidoreductase protein superfamily. A carbonic anhydrase increases the pH of the nectar to more alkaline conditions, allowing for the nonenzymatic conjugation of proline to sinapaldehyde to give nesocodin. Hydrogen peroxide is a by-product of sinapaldehyde formation that can break down the pigment; however, it is removed by a third enzyme, a catalase. The distantly related Jaltomata herrerae has red nectar and is pollinated by hummingbirds. The authors found nesocodin in the nectar of J. herrerae, as well as a carbonic anhydrase and an oxidoreductase for nesocodin biosynthesis. These findings inidicate that production of pigmented nectar in these two distantly-related vertebrate-pollinated flowers is a case of convergent evolution. (Summary by Kristen Van Gelder) PNAS 10.1073/pnas.2114420119
How many tree species are there?
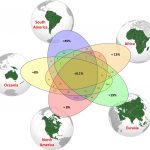
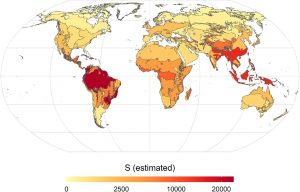 Do you know how many tree species are there? The global Forest Biodiversity Initiative (GFBI) database has annotated about 28000 tree species, while some studies indicate there could be around 60000. But why this difference? Many of the species are found in unapproachable areas. Additionally, human errors such as giving different names to the same species or considering two species to be the same, affect the ability to determine the true number of species. A recent large collaborative study by Gatti et al, with more than 100 authors, documented about 64,100 tree species. The authors estimated that there are about 9200 more tree species yet to be discovered, and that about one-third of these are likely to be rare species that need conservation. Interestingly, a large portion (about 40%) of discovered as well as undiscovered species are in the South American subcontinent. Less than 0.1% of tree species are likely present across all five continents. Identification of a true number of tree species can help in understanding the evolution of trees, their response to global environmental changes, and their biodiversity. Summary by Kamal Kumar Malukani, @KamalMalukani) PNAS, 10.1073/pnas.2115329119.
Do you know how many tree species are there? The global Forest Biodiversity Initiative (GFBI) database has annotated about 28000 tree species, while some studies indicate there could be around 60000. But why this difference? Many of the species are found in unapproachable areas. Additionally, human errors such as giving different names to the same species or considering two species to be the same, affect the ability to determine the true number of species. A recent large collaborative study by Gatti et al, with more than 100 authors, documented about 64,100 tree species. The authors estimated that there are about 9200 more tree species yet to be discovered, and that about one-third of these are likely to be rare species that need conservation. Interestingly, a large portion (about 40%) of discovered as well as undiscovered species are in the South American subcontinent. Less than 0.1% of tree species are likely present across all five continents. Identification of a true number of tree species can help in understanding the evolution of trees, their response to global environmental changes, and their biodiversity. Summary by Kamal Kumar Malukani, @KamalMalukani) PNAS, 10.1073/pnas.2115329119.