Plant Science Research Weekly: February 14
Review: Deep learning for plant genomics and crop improvement
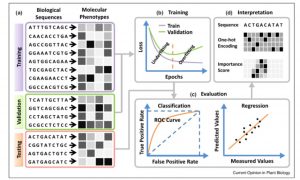 One of the goals of plant science is to use the molecular phenotype (genome, transcriptome, proteome) to predict the whole-plant phenotype. Deep learning approaches can potentially begin to do this, starting with a training dataset, and testing it with a validation dataset. Wang et al. review advances in towards this goal, particularly those that employ deep learning and neural networks, such as Convolutional Neural Networks (CNNs). The authors define different types of neural networks, and how they can be optimized for example to identify single nucleotide effects on traits, or on transcription factor binding. They also describe methods that have been developed to predict protein structure or protein-protein interactions. The authors discuss the possibilities and pitfalls of adapting deep learning models developed for animal or human genomes to plants, as well as plant-specific challenges such as polyplody. Plants’ phenotypic sensitivity to the environment adds additional complexity. The authors conclude that deep learning is a “promising approach for genome-wide identification of deleterious and adaptive variants, a prerequisite for editing-based genetic improvement of crops in future agriculture.” (Summary by Mary Williams) Curr. Opin. Plant Biol. 10.1016/j.pbi.2019.12.010
One of the goals of plant science is to use the molecular phenotype (genome, transcriptome, proteome) to predict the whole-plant phenotype. Deep learning approaches can potentially begin to do this, starting with a training dataset, and testing it with a validation dataset. Wang et al. review advances in towards this goal, particularly those that employ deep learning and neural networks, such as Convolutional Neural Networks (CNNs). The authors define different types of neural networks, and how they can be optimized for example to identify single nucleotide effects on traits, or on transcription factor binding. They also describe methods that have been developed to predict protein structure or protein-protein interactions. The authors discuss the possibilities and pitfalls of adapting deep learning models developed for animal or human genomes to plants, as well as plant-specific challenges such as polyplody. Plants’ phenotypic sensitivity to the environment adds additional complexity. The authors conclude that deep learning is a “promising approach for genome-wide identification of deleterious and adaptive variants, a prerequisite for editing-based genetic improvement of crops in future agriculture.” (Summary by Mary Williams) Curr. Opin. Plant Biol. 10.1016/j.pbi.2019.12.010
Review: Methods to visualize elements in plants
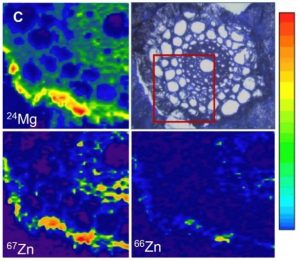 How elements are taken up, transported, accumulated, and reach homeostasis within tissues are fundamental questions in plant molecular biology. Some aspects of plant mineral nutrition and crop productivity are related to element distribution: for instance, the analysis of fertilizer translocation upon application, or how plants can tolerate high concentrations of some metals in their tissues. The distribution and concentration of nutrients in fruits and seeds are vital for human health. In a recent review, Kopittke and colleagues summarize different techniques to study element distribution in plants. In the X-ray fluorescence-based approaches, a beam of high-energy X-rays, electrons, or protons is directed to the sample, exciting different elements and allowing their identification based upon their characteristic fluorescent X-rays. In the mass spectrometry-based approaches, small portions of the sample are progressively analyzed. One of the classic techniques is autoradiography; radioactive isotopes (e.g., 26Al, 33P, 35S, 45Ca, 54Mn, 55Fe, 67Cu) can be tracked throughout the plant tissues. Another technique for in vivo analysis is the use of laser confocal microscopy with element-selective fluorophores. The currently available fluorophores allow the detection of Zn, Ni/Co, Cu, or Pb/Cd. The authors provide a comprehensive comparison of those techniques, including key features such as: accessibility, maximum resolution, scan speed, detections limits, and the suitability for in vivo analysis. (Summary by Humberto Herrera-Ubaldo) Plant Physiol. 10.1104/pp.19.01306
How elements are taken up, transported, accumulated, and reach homeostasis within tissues are fundamental questions in plant molecular biology. Some aspects of plant mineral nutrition and crop productivity are related to element distribution: for instance, the analysis of fertilizer translocation upon application, or how plants can tolerate high concentrations of some metals in their tissues. The distribution and concentration of nutrients in fruits and seeds are vital for human health. In a recent review, Kopittke and colleagues summarize different techniques to study element distribution in plants. In the X-ray fluorescence-based approaches, a beam of high-energy X-rays, electrons, or protons is directed to the sample, exciting different elements and allowing their identification based upon their characteristic fluorescent X-rays. In the mass spectrometry-based approaches, small portions of the sample are progressively analyzed. One of the classic techniques is autoradiography; radioactive isotopes (e.g., 26Al, 33P, 35S, 45Ca, 54Mn, 55Fe, 67Cu) can be tracked throughout the plant tissues. Another technique for in vivo analysis is the use of laser confocal microscopy with element-selective fluorophores. The currently available fluorophores allow the detection of Zn, Ni/Co, Cu, or Pb/Cd. The authors provide a comprehensive comparison of those techniques, including key features such as: accessibility, maximum resolution, scan speed, detections limits, and the suitability for in vivo analysis. (Summary by Humberto Herrera-Ubaldo) Plant Physiol. 10.1104/pp.19.01306
Review. Flowering plants in the Anthropocene: A political agenda
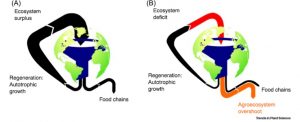 For the past 100 million years, flowering plants have been the main producers of land biomass. They also are indirectly responsible for the origin of agriculture, setting the ball rolling for the onset of the Anthropocene. Negrutiu et al. argue that the accompanying rapid global changes require a reconsideration of the relationship between humans and Earth Systems, and an a integration of planetary boundaries (biosphere integrity, water availability, global pollution, and climate change), the critical zone (interaction zone among biotic phenomena and geological Systems), and planetary health. Agriculture, which the authors describe as a “geopolitical issue,” is identified as the main stressor to these systems. In this review, the authors highlight the importance of angiosperms and the evolutionary innovations that have led to their dominance. They also describe the impacts of human intervention and breeding for “yield at any cost” and the need to consider greater nutritional quality and yield resiliency in future breeding efforts, as well as the adoption of new or underused species for food crops. Finally, the authors anticipate “the advent of a research and policy framework, integrating plant science in all sectors: the economy, local and global governance, and geopolitics.” (Summary by Mary Williams) Trends Plant Sci. 10.1016/j.tplants.2019.12.008
For the past 100 million years, flowering plants have been the main producers of land biomass. They also are indirectly responsible for the origin of agriculture, setting the ball rolling for the onset of the Anthropocene. Negrutiu et al. argue that the accompanying rapid global changes require a reconsideration of the relationship between humans and Earth Systems, and an a integration of planetary boundaries (biosphere integrity, water availability, global pollution, and climate change), the critical zone (interaction zone among biotic phenomena and geological Systems), and planetary health. Agriculture, which the authors describe as a “geopolitical issue,” is identified as the main stressor to these systems. In this review, the authors highlight the importance of angiosperms and the evolutionary innovations that have led to their dominance. They also describe the impacts of human intervention and breeding for “yield at any cost” and the need to consider greater nutritional quality and yield resiliency in future breeding efforts, as well as the adoption of new or underused species for food crops. Finally, the authors anticipate “the advent of a research and policy framework, integrating plant science in all sectors: the economy, local and global governance, and geopolitics.” (Summary by Mary Williams) Trends Plant Sci. 10.1016/j.tplants.2019.12.008
A two-way molecular dialogue between embryo and endosperm is required for seed development ($)
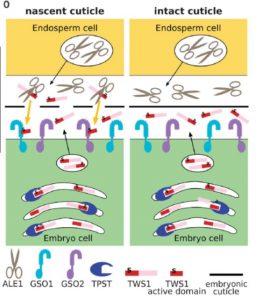 During seed formation, the developing embryo becomes covered with a water-impermeable cuticle. Several Arabidopsis mutants have been identified that are defective in cuticle formation and provide a framework for how it is formed. Doll et al. have added a missing piece to this puzzle. Briefly, embryo cells produce a secreted sulfated peptide that must be cleaved to become an active ligand for a embryonic cell-surface receptor required to promote cuticle formation. The authors show that the subtilase that cleaves the peptide precursor is produced by cells of the endosperm that surround the embryo. This suggests a model in which an incomplete cuticle allows peptide diffusion to the endosperm and back to activate cuticle formation, but when the cuticle is complete it prevents the precursor from reaching the activating subtilase, thus shutting off the cuticle-synthesis signaling. (Summary by Mary Williams) Science 10.1126/science.aaz4131
During seed formation, the developing embryo becomes covered with a water-impermeable cuticle. Several Arabidopsis mutants have been identified that are defective in cuticle formation and provide a framework for how it is formed. Doll et al. have added a missing piece to this puzzle. Briefly, embryo cells produce a secreted sulfated peptide that must be cleaved to become an active ligand for a embryonic cell-surface receptor required to promote cuticle formation. The authors show that the subtilase that cleaves the peptide precursor is produced by cells of the endosperm that surround the embryo. This suggests a model in which an incomplete cuticle allows peptide diffusion to the endosperm and back to activate cuticle formation, but when the cuticle is complete it prevents the precursor from reaching the activating subtilase, thus shutting off the cuticle-synthesis signaling. (Summary by Mary Williams) Science 10.1126/science.aaz4131
Central clock components modulate shade avoidance by directly repressing transcriptional activation activity of PIF proteins ($)
 PHYTOCHROME INTERACTING FACTORs (PIFs) are transcriptional factors that relay light signals from the phytochrome photoreceptors to regulate the expression of light-sensitive genes, but there is not a direct correlation between PIF binding and expression of the target genes. In this paper, Zhang et al. identified additional regulatory proteins that influence PIF-mediated transcription. Starting with computational predictions from ChiP-seq data, the authors found that key proteins involved in the central clock, Pseudo-Response Regulators 5 and 7 (PRR5 and PRR7) affect PIF transcriptional activity. In shade, pif mutants show reduced hypocotyl elongation, and in contrast prr mutants exhibit elongated hypocotyls, indicating an important role for these PRR clock components in light responses and hypocotyl elongation. Expression analysis of PIF-regulated genes in prr mutants showed altered gene expression suggesting PRRs repress the PIF induced gene expression. The formation of a PIF-PRR complex shows a physical interaction between PRR and PIF in the nucleus to repress PIF activity, and a direct link between the clock and light-regulated gene expression. (Summary by Suresh Damodaran) PNAS 10.1073/pnas.1918317117
PHYTOCHROME INTERACTING FACTORs (PIFs) are transcriptional factors that relay light signals from the phytochrome photoreceptors to regulate the expression of light-sensitive genes, but there is not a direct correlation between PIF binding and expression of the target genes. In this paper, Zhang et al. identified additional regulatory proteins that influence PIF-mediated transcription. Starting with computational predictions from ChiP-seq data, the authors found that key proteins involved in the central clock, Pseudo-Response Regulators 5 and 7 (PRR5 and PRR7) affect PIF transcriptional activity. In shade, pif mutants show reduced hypocotyl elongation, and in contrast prr mutants exhibit elongated hypocotyls, indicating an important role for these PRR clock components in light responses and hypocotyl elongation. Expression analysis of PIF-regulated genes in prr mutants showed altered gene expression suggesting PRRs repress the PIF induced gene expression. The formation of a PIF-PRR complex shows a physical interaction between PRR and PIF in the nucleus to repress PIF activity, and a direct link between the clock and light-regulated gene expression. (Summary by Suresh Damodaran) PNAS 10.1073/pnas.1918317117
Enhanced sustainable green revolution yield via nitrogen-responsive chromatin modulation in rice ($)
 The well-known success of the green-revolution was accomplished through the development of semi-dwarf grasses (rice and wheat), which led to decreased losses to lodging and higher yields; however, the benefits of these improved varieties depends on the application of nitrogen-rich fertilizer. As N-rich fertilizer is energetically costly and environmentally damaging, the quest is to sustain high yields but with a greater N-use efficiency. Wu et al. have identified a rice transcription factor that offers this promise. Rice plants carrying loss-of-function ngr5 (nitrogen-mediated tiller growth response 5) alleles show reduced tillering (branching) and yields, but elevated NGR5 levels (both through transgenics and in a high-yielding cultivar) was correlated with higher yields independent of N levels. The authors show that NGR5 recruits Polycomb Repressive Complex 2 to turn off tiller-inhibiting genes. Thus, the authors show “that increasing NGR5 expression or activity provides a breeding strategy to reduce nitrogen fertilizer use while boosting grain yield above what is currently sustainably achievable.” (Summary by Mary Williams) Science 10.1126/science.aaz2046
The well-known success of the green-revolution was accomplished through the development of semi-dwarf grasses (rice and wheat), which led to decreased losses to lodging and higher yields; however, the benefits of these improved varieties depends on the application of nitrogen-rich fertilizer. As N-rich fertilizer is energetically costly and environmentally damaging, the quest is to sustain high yields but with a greater N-use efficiency. Wu et al. have identified a rice transcription factor that offers this promise. Rice plants carrying loss-of-function ngr5 (nitrogen-mediated tiller growth response 5) alleles show reduced tillering (branching) and yields, but elevated NGR5 levels (both through transgenics and in a high-yielding cultivar) was correlated with higher yields independent of N levels. The authors show that NGR5 recruits Polycomb Repressive Complex 2 to turn off tiller-inhibiting genes. Thus, the authors show “that increasing NGR5 expression or activity provides a breeding strategy to reduce nitrogen fertilizer use while boosting grain yield above what is currently sustainably achievable.” (Summary by Mary Williams) Science 10.1126/science.aaz2046
DIX domain polymerization drives assembly of plant cell polarity complexes
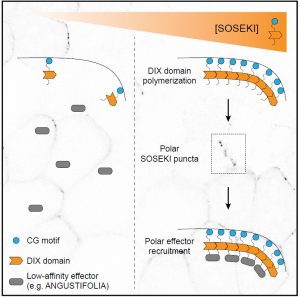 In multicellular organisms like plants, asymmetric cell division can arise from cell polarity. The signals that establish cell polarity relative to the body axis are unknown in plants. In Arabidopsis, SOSEK1 was previously identified as having a polar distribution. SOSEK1 has a DIX domain similar to the DIX oligomerization domain of the Disheveled protein involved in the establishment of polarity in animal cells. In this paper, Van Dop et al. determined the conservation and function of DIX domain-containing proteins across kingdoms. In Physcomitrella patens and Marchantia polymorpha, SOSEK1 is also localized polarly suggesting a conserved role. Structural and protein interaction analyses suggest that Arabidopsis SOSEK1 oligomerizes and this is crucial for polar localization. Pull-down assays of SOSEK1 expressed in Arabidopsis roots identified ANGUSTIFOLIA, which influences cell division. Polar localization of ANGUSTIFOLIA in Arabidopsis is altered when DIX-mediated oligomerization of SOSEK1 was hindered suggesting an important role of ANGUSTIFOLIA-SOSEK1 interaction in plant cell polarity. Thus the conservation of the DIX domain and its functional mechanism in plants is uncovered. (Summary by Suresh Damodaran) Cell 10.1016/j.cell.2020.01.011
In multicellular organisms like plants, asymmetric cell division can arise from cell polarity. The signals that establish cell polarity relative to the body axis are unknown in plants. In Arabidopsis, SOSEK1 was previously identified as having a polar distribution. SOSEK1 has a DIX domain similar to the DIX oligomerization domain of the Disheveled protein involved in the establishment of polarity in animal cells. In this paper, Van Dop et al. determined the conservation and function of DIX domain-containing proteins across kingdoms. In Physcomitrella patens and Marchantia polymorpha, SOSEK1 is also localized polarly suggesting a conserved role. Structural and protein interaction analyses suggest that Arabidopsis SOSEK1 oligomerizes and this is crucial for polar localization. Pull-down assays of SOSEK1 expressed in Arabidopsis roots identified ANGUSTIFOLIA, which influences cell division. Polar localization of ANGUSTIFOLIA in Arabidopsis is altered when DIX-mediated oligomerization of SOSEK1 was hindered suggesting an important role of ANGUSTIFOLIA-SOSEK1 interaction in plant cell polarity. Thus the conservation of the DIX domain and its functional mechanism in plants is uncovered. (Summary by Suresh Damodaran) Cell 10.1016/j.cell.2020.01.011
Rhizosphere microbiome mediates systemic root metabolite exudation ($)
 Roots exude metabolites that affect the composition and activities of their microbiome. Korenblum et al. show that the microbiome in turn affects metabolite exudation, not only locally but also systemically (shown using a split-root system). They call this response SIREM: systemically induced root exudation of metabolites. For example, the presence of Bacillus bacteria triggers exudation of various acylsugars, the composition of which is influenced by the composition of the microbiome. The authors also show a role for azelaic acid (AzA) in systemic signaling. After coating roots with thin layers of agar, the authors used MALDI-MSI to localize exuded metabolites to specific portions of the root system, for example with some derived from root hairs and others from lateral root tips; this in turn might promote distinct patterns of microbial colonization. These finding shed light on the important trans-kingdom interactions between plants and their microbiome. (Summary by Mary Williams) Proc. Natl. Acad. USA 10.1073/pnas.1912130117
Roots exude metabolites that affect the composition and activities of their microbiome. Korenblum et al. show that the microbiome in turn affects metabolite exudation, not only locally but also systemically (shown using a split-root system). They call this response SIREM: systemically induced root exudation of metabolites. For example, the presence of Bacillus bacteria triggers exudation of various acylsugars, the composition of which is influenced by the composition of the microbiome. The authors also show a role for azelaic acid (AzA) in systemic signaling. After coating roots with thin layers of agar, the authors used MALDI-MSI to localize exuded metabolites to specific portions of the root system, for example with some derived from root hairs and others from lateral root tips; this in turn might promote distinct patterns of microbial colonization. These finding shed light on the important trans-kingdom interactions between plants and their microbiome. (Summary by Mary Williams) Proc. Natl. Acad. USA 10.1073/pnas.1912130117
Farming plant cooperation in crops
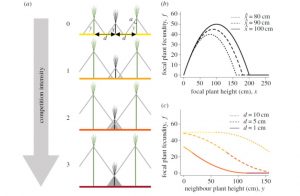 If you want a great plant, select for a strong, vigorous, high-yielding individual; this is also the outcome of natural selection. But if you want a great field of plants, these traits may not be as suitable, because the plants will expend energy competing between themselves. When seeds from many plants are used to start the next generation, there will be a mixture of tall, selfish traits and short, altruistic traits. Competitiveness has been studied in wild populations and theoretical terms, but its application to identify best practices in crops has been limited. Montazeaud et al. “develop a theoretical framework to investigate the evolution of cooperation-related traits in crops, using plant height as a case study,” and find that “combining high plant density, high relatedness and selection among groups favours the evolution of shorter plants that maximize grain yield.” They discuss their findings in terms of past and future practices, with the goal of finding low-impact approaches to high yields. (Summary by Mary Williams) Proc. R. Soc. B. 10.1098/rspb.2019.1290
If you want a great plant, select for a strong, vigorous, high-yielding individual; this is also the outcome of natural selection. But if you want a great field of plants, these traits may not be as suitable, because the plants will expend energy competing between themselves. When seeds from many plants are used to start the next generation, there will be a mixture of tall, selfish traits and short, altruistic traits. Competitiveness has been studied in wild populations and theoretical terms, but its application to identify best practices in crops has been limited. Montazeaud et al. “develop a theoretical framework to investigate the evolution of cooperation-related traits in crops, using plant height as a case study,” and find that “combining high plant density, high relatedness and selection among groups favours the evolution of shorter plants that maximize grain yield.” They discuss their findings in terms of past and future practices, with the goal of finding low-impact approaches to high yields. (Summary by Mary Williams) Proc. R. Soc. B. 10.1098/rspb.2019.1290