Plant Science Research Weekly: Feb 12, 2021
Review: On tree longevity
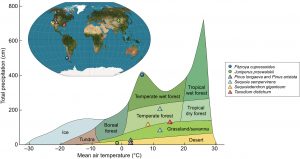 Everyone enjoys hearing about ancient trees that have been alive since various events in human history, but did you ever stop to wonder why some trees live for millennia and others don’t? If so, you’ll want to read this Tansley Review by Piovesan and Biondi. One of their first observations is that in principle, trees don’t naturally die, but instead are killed by extrinsic factors; the long-lived ones escape being killed off. A few additional observations: maximal lifespans of conifers are approximately 10x greater than angiosperms, the biggest or fastest-growing trees are almost never the oldest, very old trees are often visually distinctive with numerous dead branches, flattened tops, and partial cambial dieback resulting in bark stripping (where a tree no longer forms a complete circle). The authors address characteristics of long lived trees including the environments in which they are found, and ecological and evolutionary questions raised by them (for example, is there a selective advantage associated with extreme old age?). Finally, given the importance of climate on longevity, will long-lived trees be found in the future? This is a captivating review and one that would certainly be of interest to most students. (Summary by Mary Williams @PlantTeaching) New Phytol. 10.1111/nph.17148
Everyone enjoys hearing about ancient trees that have been alive since various events in human history, but did you ever stop to wonder why some trees live for millennia and others don’t? If so, you’ll want to read this Tansley Review by Piovesan and Biondi. One of their first observations is that in principle, trees don’t naturally die, but instead are killed by extrinsic factors; the long-lived ones escape being killed off. A few additional observations: maximal lifespans of conifers are approximately 10x greater than angiosperms, the biggest or fastest-growing trees are almost never the oldest, very old trees are often visually distinctive with numerous dead branches, flattened tops, and partial cambial dieback resulting in bark stripping (where a tree no longer forms a complete circle). The authors address characteristics of long lived trees including the environments in which they are found, and ecological and evolutionary questions raised by them (for example, is there a selective advantage associated with extreme old age?). Finally, given the importance of climate on longevity, will long-lived trees be found in the future? This is a captivating review and one that would certainly be of interest to most students. (Summary by Mary Williams @PlantTeaching) New Phytol. 10.1111/nph.17148
Genomic mechanisms of climate adaptation in polyploid bioenergy switchgrass
 Switchgrass (Panicum virgatum) is a tall, perennial species native to the prairies of North America. Switchgrass is well-known for its uses as a biofuel and forage crop. However, switchgrass is also a fascinating model to investigate how plants adapt to changing environments, as historical glacial cycles caused the switchgrass habitat to expand and contract several times. Here, Lovell et al. share a high-quality, well-annotated genome assembly for switchgrass, which is no mean feat given that this is a highly heterozygous tetraploid species. The authors sequenced 732 switchgrass genotypes and cultivated them at sites spanning over 1,800 km of latitude, from southern Texas to South Dakota. Three genetically and geographically distinct subpopulations were identified, appropriately named Midwest, Atlantic, and Gulf. Genome-wide association mapping revealed several climate–gene–biomass associations that differed between the subpopulations. Moreover, the authors discovered that during the Atlantic subpopulation’s colonization of the colder, northeastern regions of the USA, climate adaptation was augmented by the introgression of alleles from the already-adapted Midwest gene pool. This impressive study provides a wealth of genomic resources and gene–trait associations that will aid switchgrass breeding efforts. Additionally, this research provides valuable insights into how switchgrass withstands rapid environmental alterations, which will be crucial to bolstering other plant species amid climate change. (Summary by Caroline Dowling @CarolineD0wling) Nature 10.1038/s41586-020-03127-1
Switchgrass (Panicum virgatum) is a tall, perennial species native to the prairies of North America. Switchgrass is well-known for its uses as a biofuel and forage crop. However, switchgrass is also a fascinating model to investigate how plants adapt to changing environments, as historical glacial cycles caused the switchgrass habitat to expand and contract several times. Here, Lovell et al. share a high-quality, well-annotated genome assembly for switchgrass, which is no mean feat given that this is a highly heterozygous tetraploid species. The authors sequenced 732 switchgrass genotypes and cultivated them at sites spanning over 1,800 km of latitude, from southern Texas to South Dakota. Three genetically and geographically distinct subpopulations were identified, appropriately named Midwest, Atlantic, and Gulf. Genome-wide association mapping revealed several climate–gene–biomass associations that differed between the subpopulations. Moreover, the authors discovered that during the Atlantic subpopulation’s colonization of the colder, northeastern regions of the USA, climate adaptation was augmented by the introgression of alleles from the already-adapted Midwest gene pool. This impressive study provides a wealth of genomic resources and gene–trait associations that will aid switchgrass breeding efforts. Additionally, this research provides valuable insights into how switchgrass withstands rapid environmental alterations, which will be crucial to bolstering other plant species amid climate change. (Summary by Caroline Dowling @CarolineD0wling) Nature 10.1038/s41586-020-03127-1
Regulation of ACD6 ion channel-like protein by small peptides
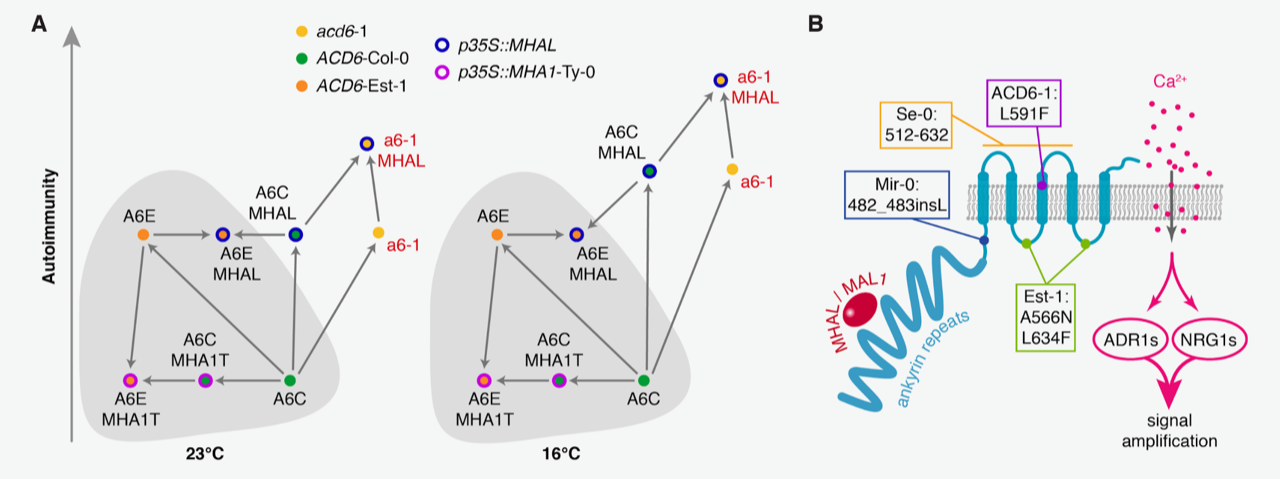 ACCELERATED CELL DEATH 6 (ACD6) positively regulates the signaling of the plant defense hormone salicylic acid (SA). It was originally identified by the study of natural variation in defense responses of Arabidopsis thaliana. The naturally hyperactive ACD6-Est-I allele confers broad-spectrum disease resistance at the cost of reduced growth due to constitutive activation of SA-mediated immunity. Yet, not all ACD6-Est-I carriers show such “autoimmunity,” suggesting that ACD6-Est-I-mediated immune responses can be suppressed by epistatic interactions. Here, through genome-wide association (GWA) analysis focusing on ACD6-Est-I carriers, Zhu et al. identified MODIFIER OF HYPERACTIVE ACD6 I (MHAI) as a suppressor of ACD6-mediated immunity. MHAI and its paralog MHAI-LIKE (MHAL) encode small peptides that interact with ACD6 protein in planta, and when overexpressed, MHAL enhanced activity of the non-autoimmune allele of ACD6. The authors found that ACD6 has structurally similar domains to mammalian transient receptor potential channels, and surprisingly, they found that ACD6 stimulates Ca2+ influx when expressed in oocytes. The potential ion channel activity of ACD6 was enhanced by MHAL. Finally, the authors showed that ACD6 triggered autoimmunity requires Ca2+. This study reveals complicated epistatic interactions between a key immune regulator and previously unidentified small peptides by leveraging the power of GWA analysis. (Summary by Tatsuya Nobori @nobolly) bioRxiv 10.1101/2021.01.25.428077
ACCELERATED CELL DEATH 6 (ACD6) positively regulates the signaling of the plant defense hormone salicylic acid (SA). It was originally identified by the study of natural variation in defense responses of Arabidopsis thaliana. The naturally hyperactive ACD6-Est-I allele confers broad-spectrum disease resistance at the cost of reduced growth due to constitutive activation of SA-mediated immunity. Yet, not all ACD6-Est-I carriers show such “autoimmunity,” suggesting that ACD6-Est-I-mediated immune responses can be suppressed by epistatic interactions. Here, through genome-wide association (GWA) analysis focusing on ACD6-Est-I carriers, Zhu et al. identified MODIFIER OF HYPERACTIVE ACD6 I (MHAI) as a suppressor of ACD6-mediated immunity. MHAI and its paralog MHAI-LIKE (MHAL) encode small peptides that interact with ACD6 protein in planta, and when overexpressed, MHAL enhanced activity of the non-autoimmune allele of ACD6. The authors found that ACD6 has structurally similar domains to mammalian transient receptor potential channels, and surprisingly, they found that ACD6 stimulates Ca2+ influx when expressed in oocytes. The potential ion channel activity of ACD6 was enhanced by MHAL. Finally, the authors showed that ACD6 triggered autoimmunity requires Ca2+. This study reveals complicated epistatic interactions between a key immune regulator and previously unidentified small peptides by leveraging the power of GWA analysis. (Summary by Tatsuya Nobori @nobolly) bioRxiv 10.1101/2021.01.25.428077
A stable antimicrobial peptide with dual functions of treating and preventing citrus Huanglongbing
 Citrus Huanglongbing (HLB) caused by the bacterium Candidatus Liberibacter asiaticus (CLas) is the most destructive disease of citrus and currently has no cure. Current management practices are also not effective. Huang et al. used comparative analysis of small RNAs and mRNAs between HLB-sensitive and HLB-tolerant hybrids to identify genes mediating HLB tolerance. One of these genes encodes a 67 amino acid peptide that has an Arabidopsis homolog with antimicrobial properties. This candidate gene was subsequently named SAMP; stable antimicrobial peptide. To functionally characterize this candidate gene, SAMP genes were cloned from HLB tolerant citrus relatives. Tolerant citrus relatives have both a long (109-aa) and short (67-aa) version of the peptide, while the susceptible varieties have only the long version (LSAMP). SAMP mRNA expression was significantly higher in tolerant varieties than LSAMP expression in the susceptible ones. An antibody was used to show that the 6.7 kD short version was present in the phloem of tolerant varieties but not in the susceptible ones. SAMP-containing solutions was also shown to supress CLas in HLB-positive citrus trees and protected healthy trees against infection by activating defense response. The authors also showed that SAMP is heat stable and effective against α-proteobacteria by causing rapid cytosol leakage and lysis. Finally, several truncated versions of SAMP showed that the second α-helix of SAMP is responsible for the antibacterial activity of SAMP. (Summary by Toluwase Olukayode @toluxylic) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2019628118
Citrus Huanglongbing (HLB) caused by the bacterium Candidatus Liberibacter asiaticus (CLas) is the most destructive disease of citrus and currently has no cure. Current management practices are also not effective. Huang et al. used comparative analysis of small RNAs and mRNAs between HLB-sensitive and HLB-tolerant hybrids to identify genes mediating HLB tolerance. One of these genes encodes a 67 amino acid peptide that has an Arabidopsis homolog with antimicrobial properties. This candidate gene was subsequently named SAMP; stable antimicrobial peptide. To functionally characterize this candidate gene, SAMP genes were cloned from HLB tolerant citrus relatives. Tolerant citrus relatives have both a long (109-aa) and short (67-aa) version of the peptide, while the susceptible varieties have only the long version (LSAMP). SAMP mRNA expression was significantly higher in tolerant varieties than LSAMP expression in the susceptible ones. An antibody was used to show that the 6.7 kD short version was present in the phloem of tolerant varieties but not in the susceptible ones. SAMP-containing solutions was also shown to supress CLas in HLB-positive citrus trees and protected healthy trees against infection by activating defense response. The authors also showed that SAMP is heat stable and effective against α-proteobacteria by causing rapid cytosol leakage and lysis. Finally, several truncated versions of SAMP showed that the second α-helix of SAMP is responsible for the antibacterial activity of SAMP. (Summary by Toluwase Olukayode @toluxylic) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2019628118
Rhizobia use a pathogenic-like effector to hijack leguminous nodulation signaling
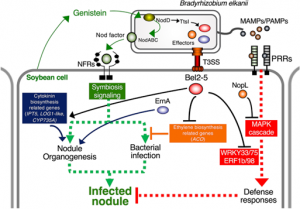 Symbiotic interactions between legume plants and compatible rhizobia bacteria lead to the formation of nitrogen-fixing nodules in the host plant root. Compatibility between rhizobia and host plants is determined by various factors like plant species-specific flavonoid secretion, extracellular polysaccharides of bacteria, and the effector proteins secreted by the rhizobia type III secretion system. These effector proteins were initially identified in pathogenic bacteria as involved in pathogenicity and bacterial infection. Various studies have shown that these effector proteins are induced by host flavonoids and are involved in activating nodulation in the absence of common symbiosis signaling pathway, nod factor (NF) signaling. To understand the mechanism of effector protein during symbiosis between the rhizobia USDA61 and soybean plant, Ratu et al. identified Bel2-5, an effector protein playing a prominent role in promoting nodulation in the nfr1 soybean mutant. Phylogenetic and in-silico analysis of its protein sequence showed similarity with the effector protein from pathogenic bacteria Xanthomonas campestris. Further analysis of the conserved domain at its C terminus, the ULP1 like domain, showed its vital role in nodule formation. Additionally, soybean root transcriptome analysis showed the involvement of Bel2-5 in inducing cytokinin related genes and repressing defense related genes, both of which are essential for nodulation. Overall, the authors showed that rhizobia bacteria use pathogenic effectors to hijack the common symbiosis signaling pathway and promote nodulation. (Summary by Sunita Pathak @psunita980) Sci. Reports 10.1038/s41598-021-81598-6
Symbiotic interactions between legume plants and compatible rhizobia bacteria lead to the formation of nitrogen-fixing nodules in the host plant root. Compatibility between rhizobia and host plants is determined by various factors like plant species-specific flavonoid secretion, extracellular polysaccharides of bacteria, and the effector proteins secreted by the rhizobia type III secretion system. These effector proteins were initially identified in pathogenic bacteria as involved in pathogenicity and bacterial infection. Various studies have shown that these effector proteins are induced by host flavonoids and are involved in activating nodulation in the absence of common symbiosis signaling pathway, nod factor (NF) signaling. To understand the mechanism of effector protein during symbiosis between the rhizobia USDA61 and soybean plant, Ratu et al. identified Bel2-5, an effector protein playing a prominent role in promoting nodulation in the nfr1 soybean mutant. Phylogenetic and in-silico analysis of its protein sequence showed similarity with the effector protein from pathogenic bacteria Xanthomonas campestris. Further analysis of the conserved domain at its C terminus, the ULP1 like domain, showed its vital role in nodule formation. Additionally, soybean root transcriptome analysis showed the involvement of Bel2-5 in inducing cytokinin related genes and repressing defense related genes, both of which are essential for nodulation. Overall, the authors showed that rhizobia bacteria use pathogenic effectors to hijack the common symbiosis signaling pathway and promote nodulation. (Summary by Sunita Pathak @psunita980) Sci. Reports 10.1038/s41598-021-81598-6
When fungi get ‘SCOOP’ed: MIK2 receptor kinase perceives SCOOP phytocytokines in Arabidopsis thaliana
 Plant cells sense endogenous and exogenous molecules through proteins localized to the cell surface. While numerous ligands that mediate a variety of developmental and stress processes are known, cognate receptors for many ligands remain unidentified. Rhodes and colleagues have now shown the Arabidopsis thaliana leucine-rich receptor kinase MIK2 to be the receptor for the SCOOP (SERINE RICH ENDOGENOUS PEPTIDE) family of secreted peptides that function as ‘phytocytokines” (in mammals, cytokines are immune-regulating secreted peptides). Testing a range of microbe and plant-derived elicitors, the authors found mik2 mutants to be unresponsive to SCOOP peptides. They found SCOOP peptides directly bind MIK2 protein and induce MIK2 binding with its co-receptor, BAK1, as well as BAK1 phosphorylation. Significantly, peptides with SCOOP-like motifs from the pathogenic fungi belonging to Fusarium spp. also induced responses similar to Arabidopsis SCOOP peptides, illuminating the basis for MIK2-based resistance fungal pathogens. (Summary by Pavithran Narayanan @pavi_narayanan) Nature Comms. 10.1038/s41467-021-20932-y.
Plant cells sense endogenous and exogenous molecules through proteins localized to the cell surface. While numerous ligands that mediate a variety of developmental and stress processes are known, cognate receptors for many ligands remain unidentified. Rhodes and colleagues have now shown the Arabidopsis thaliana leucine-rich receptor kinase MIK2 to be the receptor for the SCOOP (SERINE RICH ENDOGENOUS PEPTIDE) family of secreted peptides that function as ‘phytocytokines” (in mammals, cytokines are immune-regulating secreted peptides). Testing a range of microbe and plant-derived elicitors, the authors found mik2 mutants to be unresponsive to SCOOP peptides. They found SCOOP peptides directly bind MIK2 protein and induce MIK2 binding with its co-receptor, BAK1, as well as BAK1 phosphorylation. Significantly, peptides with SCOOP-like motifs from the pathogenic fungi belonging to Fusarium spp. also induced responses similar to Arabidopsis SCOOP peptides, illuminating the basis for MIK2-based resistance fungal pathogens. (Summary by Pavithran Narayanan @pavi_narayanan) Nature Comms. 10.1038/s41467-021-20932-y.
Chromatin phosphoproteomics identifies an AT-hook motif protein involved in PAMP-triggered immunity
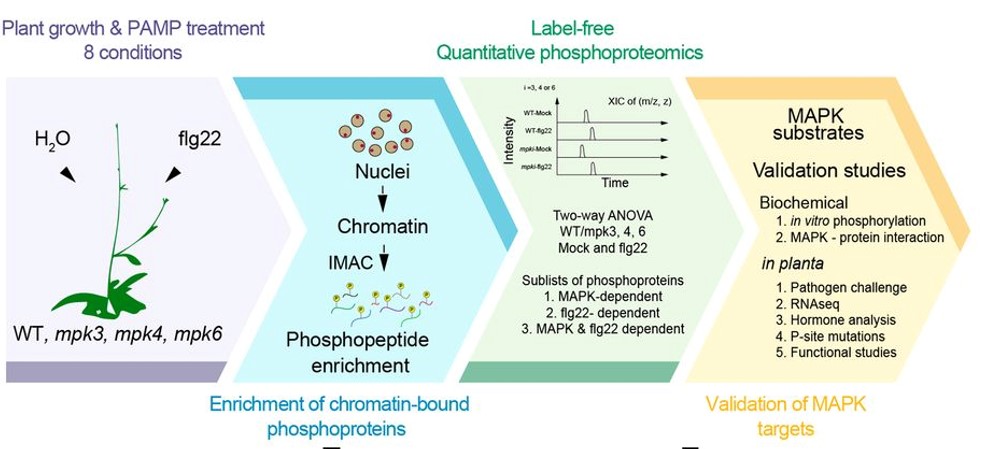 Mitogen-activated protein kinases (MAPKs) are conserved protein kinases in eukaryotes that participate in signaling from cytoplasmic to chromatin events to allow transcription reprogramming. MAPKs play prominent roles at the chromatin level. Rayapuram et al. report a chromatin-associated phosphoproteome from wild type Arabidopsis and mpk3/4/6 mutants, with and without a 15-min treatment with flg22. They identified 56 differentially phosphorylated peptides of which 38 possess a MAPK phosphorylation motif including factors involved in chromatin organization, DNA binding, RNA processing and splicing. Two of the sites are on the AT-hook motif nuclear localized protein 13 (AHL13), a DNA-binding transcription factor. AHL13 preferentially interacts with MPK6 and is phosphorylated by MPK3/4/6. It is involved in regulating disease resistance to bacteria, with the mutant and phosphodead form more susceptible to pathogens. Transcriptome analysis revealed that AHL13 regulates jasmonic acid biosynthesis and signaling genes. Interestingly, the phosphodead form is degraded by the proteasome-dependent pathway and the plant becomes more sensitive to pathogens. This study sheds light on identification a set of phosphorylated chromatin substrates that regulate plant immunity. (Summary by Min May Wong @wongminmay) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2004670118
Mitogen-activated protein kinases (MAPKs) are conserved protein kinases in eukaryotes that participate in signaling from cytoplasmic to chromatin events to allow transcription reprogramming. MAPKs play prominent roles at the chromatin level. Rayapuram et al. report a chromatin-associated phosphoproteome from wild type Arabidopsis and mpk3/4/6 mutants, with and without a 15-min treatment with flg22. They identified 56 differentially phosphorylated peptides of which 38 possess a MAPK phosphorylation motif including factors involved in chromatin organization, DNA binding, RNA processing and splicing. Two of the sites are on the AT-hook motif nuclear localized protein 13 (AHL13), a DNA-binding transcription factor. AHL13 preferentially interacts with MPK6 and is phosphorylated by MPK3/4/6. It is involved in regulating disease resistance to bacteria, with the mutant and phosphodead form more susceptible to pathogens. Transcriptome analysis revealed that AHL13 regulates jasmonic acid biosynthesis and signaling genes. Interestingly, the phosphodead form is degraded by the proteasome-dependent pathway and the plant becomes more sensitive to pathogens. This study sheds light on identification a set of phosphorylated chromatin substrates that regulate plant immunity. (Summary by Min May Wong @wongminmay) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2004670118



