Plant Science Research Weekly: December 2, 2022
Review: The power and perils of de novo domestication using gene editing
 I’m excited by the ways that knowledge about plant developmental and metabolic programs is being used to make new varieties of plants. This excellent review describes some of these applications such as changes in fruit size or seed number. De novo domestication is a specific type of application that allows the introduction of some of these desired traits (selected over thousands of years) into wild, undomesticated plants, and there are many approaches being employed. Traits that facilitate breeding, for example by rendering plants self-compatible or introducing monoecy (separate male and female flowers on a single plant, as in corn) can be introduced to support further domestication efforts. But which plants to start with? Unlike botanists of yore, we now recognize, respect, and acknowledge the contributions of indigenous knowledge into our discipline. As written here, “Working with indigenous knowledge holders should be collaborative, not extractive….. Technological innovation that grounds local contexts (economic, social, ecological) and that includes scientists and farmers from the Global South from the very beginning will go a long way to prevent history from repeating itself.” Bravo! (Summary by Mary Williams @PlantTeaching) Annu. Rev. Plant Biol. 10.1146/annurev-arplant-053122-030653
I’m excited by the ways that knowledge about plant developmental and metabolic programs is being used to make new varieties of plants. This excellent review describes some of these applications such as changes in fruit size or seed number. De novo domestication is a specific type of application that allows the introduction of some of these desired traits (selected over thousands of years) into wild, undomesticated plants, and there are many approaches being employed. Traits that facilitate breeding, for example by rendering plants self-compatible or introducing monoecy (separate male and female flowers on a single plant, as in corn) can be introduced to support further domestication efforts. But which plants to start with? Unlike botanists of yore, we now recognize, respect, and acknowledge the contributions of indigenous knowledge into our discipline. As written here, “Working with indigenous knowledge holders should be collaborative, not extractive….. Technological innovation that grounds local contexts (economic, social, ecological) and that includes scientists and farmers from the Global South from the very beginning will go a long way to prevent history from repeating itself.” Bravo! (Summary by Mary Williams @PlantTeaching) Annu. Rev. Plant Biol. 10.1146/annurev-arplant-053122-030653
Time to update the nomenclature of plant ribosomal proteins
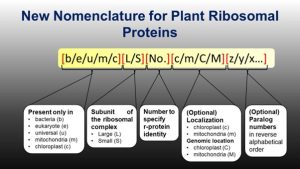 The ribosome is a macromolecular structure made up of many subunits. The proteins in these subunits were named based on their sedimentation rates and gel mobility, but different groups gave the proteins different names in various species, not based on homology. A ribosomal protein (r-protein) nomenclature system was proposed that indicates the origin of the r-protein (e.g., b for bacteria, e for eukaryote, etc.), but it is not adequate for plants as, unlike animals, plant organelle genomes also encode r-proteins, and plants carry multiple genes encoding the same r-proteins. Now Scarpin et al. propose an upgrade to the system of plant r-protein nomenclature. Specifically, they propose to (1) add a suffix, ‘c’ or ‘m’, for chloroplast and mitochondrial-targeted r-proteins, (2) capitalize ‘C’ or ‘M’ if the gene encoding that protein is encoded in respective organelle genome and (3) add a suffix starting from the end of alphabet (z, y, x and so on) for paralogs. These upgrades are proposed based on feedback received on a preprint, social media, email communications, and discussions at the Plant Biology 2022 conference. (Summary by Kamal Kumar Malukani, @KamalMalukani) Plant Cell 10.1093/plcell/koac333
The ribosome is a macromolecular structure made up of many subunits. The proteins in these subunits were named based on their sedimentation rates and gel mobility, but different groups gave the proteins different names in various species, not based on homology. A ribosomal protein (r-protein) nomenclature system was proposed that indicates the origin of the r-protein (e.g., b for bacteria, e for eukaryote, etc.), but it is not adequate for plants as, unlike animals, plant organelle genomes also encode r-proteins, and plants carry multiple genes encoding the same r-proteins. Now Scarpin et al. propose an upgrade to the system of plant r-protein nomenclature. Specifically, they propose to (1) add a suffix, ‘c’ or ‘m’, for chloroplast and mitochondrial-targeted r-proteins, (2) capitalize ‘C’ or ‘M’ if the gene encoding that protein is encoded in respective organelle genome and (3) add a suffix starting from the end of alphabet (z, y, x and so on) for paralogs. These upgrades are proposed based on feedback received on a preprint, social media, email communications, and discussions at the Plant Biology 2022 conference. (Summary by Kamal Kumar Malukani, @KamalMalukani) Plant Cell 10.1093/plcell/koac333
Structure of a TOC-TIC supercomplex spanning two chloroplast envelope membranes
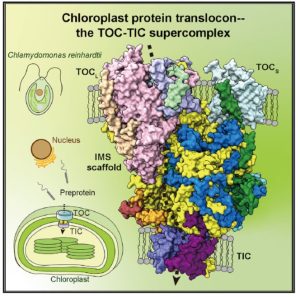 In an evolutionary plot twist, most of the proteins needed inside the chloroplast are encoded in the nucleus and translated in the cytosol as preproteins. The preproteins are imported into the chloroplast through two membranes (outer and inner). Genetic and biochemical approaches have revealed many of the components of the multi-protein complexes that allow this import, known as TOC (translocon outer membrane chloroplast) and TIC (translocon inner membrane chloroplast; the phylogenetically distant structures with similar function in mitochondria are known as TOM and TIM). Now, the awesome power of cryo-EM has revealed the structures of these complexes from Chlamydomonas in outstanding detail, providing new insights into how protein import is conducted. The work reveals that the membrane-spanning channel of TOC is made up of β-barrel pores whereas that of TIC mainly consists of transmembrane helices. Furthermore, TOC and TIC are held together into a supercomplex by an intermembrane space (IMS) scaffold that has a hydrophilic cleft that likely guides the translocation of the preprotein. The authors note that some of the proteins that make up the TOC-TIC supercomplex are restricted to certain lineages; for example, the IMS scaffold components are absent in some Poaceae species including wheat, rice and maize, raising questions of what this structure looks like in those species. (Summary by Mary Williams @PlantTeaching) Cell 10.1016/j.cell.2022.10.030
In an evolutionary plot twist, most of the proteins needed inside the chloroplast are encoded in the nucleus and translated in the cytosol as preproteins. The preproteins are imported into the chloroplast through two membranes (outer and inner). Genetic and biochemical approaches have revealed many of the components of the multi-protein complexes that allow this import, known as TOC (translocon outer membrane chloroplast) and TIC (translocon inner membrane chloroplast; the phylogenetically distant structures with similar function in mitochondria are known as TOM and TIM). Now, the awesome power of cryo-EM has revealed the structures of these complexes from Chlamydomonas in outstanding detail, providing new insights into how protein import is conducted. The work reveals that the membrane-spanning channel of TOC is made up of β-barrel pores whereas that of TIC mainly consists of transmembrane helices. Furthermore, TOC and TIC are held together into a supercomplex by an intermembrane space (IMS) scaffold that has a hydrophilic cleft that likely guides the translocation of the preprotein. The authors note that some of the proteins that make up the TOC-TIC supercomplex are restricted to certain lineages; for example, the IMS scaffold components are absent in some Poaceae species including wheat, rice and maize, raising questions of what this structure looks like in those species. (Summary by Mary Williams @PlantTeaching) Cell 10.1016/j.cell.2022.10.030
THP9 enhances seed protein content and nitrogen-use efficiency in maize
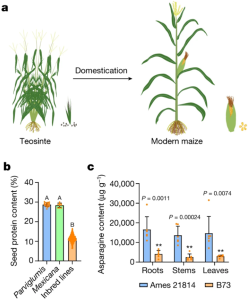 Increasing nitrogen-use efficiency (NUE) and seed protein content are important for maize breeding. Modern maize hybrids have 6.5-16.5% seed protein content, while the wild ancestor of maize, called teosinte, has ~30% seed protein content. In addition, teosinte (accession Ames 21814) contains notably higher levels of asparagine than modern maize (accession B73). Huang et al. crossed Ames 21814 with B73 to perform genome assembly and map-based cloning, from which they identified an important high-protein quantitative trait locus (QTL), named TEOSINTE HIGH PROTEIN9 (THP9). The THP9 QTL contains an asparagine synthetase 4 (ASN4) gene. The B73 THP9 locus contains a premature stop codon, making it a null allele. Overexpressing teosinte THP9 in B73 increased the seed protein content from ~12% to 14%-17%, and improved NUE in low-nitrogen conditions. The identification of THP9 will benefit maize breeding, which could potentially reduce the use of nitrogen fertilizers and produce more vegetable proteins for human diets. Notably, the overexpression of teosinte THP9 locus in B73 did not fully increase the seed protein content level to teosinte level, indicating there are more high-protein QTLs to be discovered. (Summary by Xiaohui Li @Xiao_hui_Li) Nature 10.1038/s41586-022-05441-2
Increasing nitrogen-use efficiency (NUE) and seed protein content are important for maize breeding. Modern maize hybrids have 6.5-16.5% seed protein content, while the wild ancestor of maize, called teosinte, has ~30% seed protein content. In addition, teosinte (accession Ames 21814) contains notably higher levels of asparagine than modern maize (accession B73). Huang et al. crossed Ames 21814 with B73 to perform genome assembly and map-based cloning, from which they identified an important high-protein quantitative trait locus (QTL), named TEOSINTE HIGH PROTEIN9 (THP9). The THP9 QTL contains an asparagine synthetase 4 (ASN4) gene. The B73 THP9 locus contains a premature stop codon, making it a null allele. Overexpressing teosinte THP9 in B73 increased the seed protein content from ~12% to 14%-17%, and improved NUE in low-nitrogen conditions. The identification of THP9 will benefit maize breeding, which could potentially reduce the use of nitrogen fertilizers and produce more vegetable proteins for human diets. Notably, the overexpression of teosinte THP9 locus in B73 did not fully increase the seed protein content level to teosinte level, indicating there are more high-protein QTLs to be discovered. (Summary by Xiaohui Li @Xiao_hui_Li) Nature 10.1038/s41586-022-05441-2
Hydraulic flux-responsive hormone redistribution determines root branching
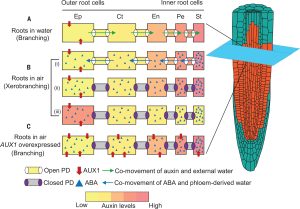 Climate change affects rain patterns and hydrological cycles all over the world, exacerbating water scarcity issues. Root branching is altered by local spatial differences in soil moisture. Indeed, when root tips temporarily lose contact with moisture, root branching stops until the appropriate moisture level is encountered again. This response is named xerobranching and in this work, Mehra et al. decipher its mechanistic basis. They prove, both genetically and with a FRET biosensor, that ABA represses lateral root development. During xerobranching response, root-tip tissues rely on an outward flux of phloem-derived water (in which ABA travels) to maintain growth. By using a fluorescent tracer, the authors demonstrated that the water flow travels through plasmodesmata, which close when they perceive ABA. The closure of plasmodesmata does not allow the required hormone auxin to travel inwards, and thus lateral roots cannot be developed. The fact that climate change is going to affect how water is distributed in the soil strengthens the need to develop resilient crops under these conditions. (Summary by Eva Maria Gomez Alvarez, @eva_ga96) Science 10.1126/science.add3771
Climate change affects rain patterns and hydrological cycles all over the world, exacerbating water scarcity issues. Root branching is altered by local spatial differences in soil moisture. Indeed, when root tips temporarily lose contact with moisture, root branching stops until the appropriate moisture level is encountered again. This response is named xerobranching and in this work, Mehra et al. decipher its mechanistic basis. They prove, both genetically and with a FRET biosensor, that ABA represses lateral root development. During xerobranching response, root-tip tissues rely on an outward flux of phloem-derived water (in which ABA travels) to maintain growth. By using a fluorescent tracer, the authors demonstrated that the water flow travels through plasmodesmata, which close when they perceive ABA. The closure of plasmodesmata does not allow the required hormone auxin to travel inwards, and thus lateral roots cannot be developed. The fact that climate change is going to affect how water is distributed in the soil strengthens the need to develop resilient crops under these conditions. (Summary by Eva Maria Gomez Alvarez, @eva_ga96) Science 10.1126/science.add3771
TTL proteins identified as a new direct interactor of both cellulose synthase complexes and microtubules
 Cellulose synthase (CESA) complexes (CSCs) synthesize the main polysaccharide component of plant primary cell wall, cellulose. The trafficking and dynamics of CSC are tightly regulated. Kesten et al. identified a new family of CSC- and microtubule-interacting proteins, named TETRATRICOPEPTIDE THIOREDOXIN-LIKE (TTL) proteins, which help to maintain cellulose synthesis under salt stress. The ttl1 ttl3 double mutant is hypersensitive to salt stress, and contains less cellulose content comparing to wild type under salt stress. The CSC foci density decreases more rapidly in ttl1 ttl3 double mutant upon salt stress, and fails to recover in 32 h, compared to wild type. And interestingly, TTL3 predominantly localizes to cytosolic regions under control conditions, but re-localizes to the plasma membrane (PM) upon salt stress, revealed by live-cell imaging and subcellular fractionation analysis. Additionally, TTL3 directly interacts with the cytosolic catalytic domain of CESA1. Live-cell imaging provides evidence that TTL3-GFP and tdTomato-CESA6 colocalize and comigrate at the PM, suggesting that the TTL protein might be a component of CSC. Furthermore, TTL3 can directly interact with microtubules. Taken together, TTL proteins might bridge salt stress perception with the dynamic control of cellulose synthesis at the PM. (Summary by Xiaohui Li @Xiao_hui_Li) Sci. Adv. 10.1126/sciadv.abq6971
Cellulose synthase (CESA) complexes (CSCs) synthesize the main polysaccharide component of plant primary cell wall, cellulose. The trafficking and dynamics of CSC are tightly regulated. Kesten et al. identified a new family of CSC- and microtubule-interacting proteins, named TETRATRICOPEPTIDE THIOREDOXIN-LIKE (TTL) proteins, which help to maintain cellulose synthesis under salt stress. The ttl1 ttl3 double mutant is hypersensitive to salt stress, and contains less cellulose content comparing to wild type under salt stress. The CSC foci density decreases more rapidly in ttl1 ttl3 double mutant upon salt stress, and fails to recover in 32 h, compared to wild type. And interestingly, TTL3 predominantly localizes to cytosolic regions under control conditions, but re-localizes to the plasma membrane (PM) upon salt stress, revealed by live-cell imaging and subcellular fractionation analysis. Additionally, TTL3 directly interacts with the cytosolic catalytic domain of CESA1. Live-cell imaging provides evidence that TTL3-GFP and tdTomato-CESA6 colocalize and comigrate at the PM, suggesting that the TTL protein might be a component of CSC. Furthermore, TTL3 can directly interact with microtubules. Taken together, TTL proteins might bridge salt stress perception with the dynamic control of cellulose synthesis at the PM. (Summary by Xiaohui Li @Xiao_hui_Li) Sci. Adv. 10.1126/sciadv.abq6971
ROP signaling regulates spatial pattern of cell division and specification of meristem notch
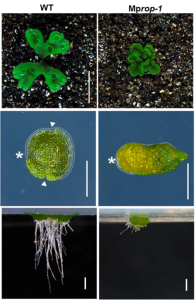 Precise control of cell division is an important requirement for proper development in multicellular organisms. Rho-like GTPases from Plants (ROPs) are key conserved regulators of cell polarity and morphogenesis, however, it is unknown if ROP signaling pathways regulate cell division patterning and meristem formation during embryogenesis and organogenesis. Additionally, the high degree of genetic redundancy in angiosperms (Arabidopsis encodes 11 ROPs) has hindered ROP functional studies. The bryophyte Marchantia polymorpha encodes a single ROP gene (MpROP), which makes it an excellent system to explore ROP functions during development, such as gemma formation, a process like embryogenesis or axillary meristem formation in angiosperms. Rong et al. show that MpROP localizes at the tip of growing rhizoids, and Mprop loss-of-function mutants display defective rhizoid growth, indicating an evolutionary conserved ROP function during polar tip growth in land plants. Strikingly, Mprop mutant gemmae display an absence of meristem notches and abnormal cell division patterns in the thalli and meristem notch-like area as well as reduced anisotropy of microtubules, indicating that MpROP likely regulates cortical microtubules organization, which in turns impacts the position and orientation of cell division planes. Finally, the authors were able to rescue the defective phenotypes observed in Mprop mutants by complementation using Arabidopsis ROP2, indicating that ROPs functions are broadly conserved among land plants. This interesting paper effectively establishes and validates Marchantia as a promising and simple system to elucidate the function of ROP proteins in complex developmental processes such as meristem formation and cell division spatial patterning. (Summary by Jesus Leon @jesussaur) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2117803119
Precise control of cell division is an important requirement for proper development in multicellular organisms. Rho-like GTPases from Plants (ROPs) are key conserved regulators of cell polarity and morphogenesis, however, it is unknown if ROP signaling pathways regulate cell division patterning and meristem formation during embryogenesis and organogenesis. Additionally, the high degree of genetic redundancy in angiosperms (Arabidopsis encodes 11 ROPs) has hindered ROP functional studies. The bryophyte Marchantia polymorpha encodes a single ROP gene (MpROP), which makes it an excellent system to explore ROP functions during development, such as gemma formation, a process like embryogenesis or axillary meristem formation in angiosperms. Rong et al. show that MpROP localizes at the tip of growing rhizoids, and Mprop loss-of-function mutants display defective rhizoid growth, indicating an evolutionary conserved ROP function during polar tip growth in land plants. Strikingly, Mprop mutant gemmae display an absence of meristem notches and abnormal cell division patterns in the thalli and meristem notch-like area as well as reduced anisotropy of microtubules, indicating that MpROP likely regulates cortical microtubules organization, which in turns impacts the position and orientation of cell division planes. Finally, the authors were able to rescue the defective phenotypes observed in Mprop mutants by complementation using Arabidopsis ROP2, indicating that ROPs functions are broadly conserved among land plants. This interesting paper effectively establishes and validates Marchantia as a promising and simple system to elucidate the function of ROP proteins in complex developmental processes such as meristem formation and cell division spatial patterning. (Summary by Jesus Leon @jesussaur) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2117803119
Cuticle chemistry drives the development of diffraction gratings on the surface of Hibiscus trionum petals
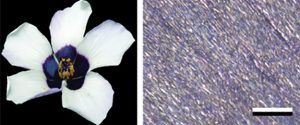 The long, long history of plant-pollinator interactions has led to a stunning variety of flower forms, including variations in petal color and iridescence. Color is caused by the accumulation of various pigments that selectively absorb certain wavelengths of light, whereas iridescence arises from surface structures that selectively diffract light (see Physics of Pollinator Attraction). In particular, diffraction gratings are ordered systems of ridges and grooves that allow constructive interference for different reflected wavelengths at different angles. In this work, Moyroud et al. explore the origin of these diffractive gratings, using a new model system of Hibiscus flowers. Specifically, they followed petal development over time to identify when these cuticular striation patterns arise. They found that cuticular striations can be independent of cuticle thickness or cell shape, but are affected by cuticle composition or the arrangement of its constituents, which may affect its material properties and therefore susceptibility to form striations. (Summary by Mary Williams @PlantTeaching) Curr. Biol. 10.1016/j.cub.2022.10.065
The long, long history of plant-pollinator interactions has led to a stunning variety of flower forms, including variations in petal color and iridescence. Color is caused by the accumulation of various pigments that selectively absorb certain wavelengths of light, whereas iridescence arises from surface structures that selectively diffract light (see Physics of Pollinator Attraction). In particular, diffraction gratings are ordered systems of ridges and grooves that allow constructive interference for different reflected wavelengths at different angles. In this work, Moyroud et al. explore the origin of these diffractive gratings, using a new model system of Hibiscus flowers. Specifically, they followed petal development over time to identify when these cuticular striation patterns arise. They found that cuticular striations can be independent of cuticle thickness or cell shape, but are affected by cuticle composition or the arrangement of its constituents, which may affect its material properties and therefore susceptibility to form striations. (Summary by Mary Williams @PlantTeaching) Curr. Biol. 10.1016/j.cub.2022.10.065
Calcium-mediated rapid movements defend against herbivorous insects in Mimosa pudica
 Plants that respond to touch, like Mimosa pudica (aka the sensitive plant) are great ways to introduce children to the fascination of plants. If you haven’t had the chance to explore this plant, a touch to a leaflet causes the leaflets to fold into themselves, and recurring touches cause the leaf to drop away. Both responses occur due to a rapid loss of water from critical cells at hinge points. Hagihara et al. introduced into M. pudica an ultrasensitive fluorescent calcium ion (Ca2+) indicator (GCaMP6f) to investigate these fast responses. Wounding elicited a rapid increase in cytosolic calcium that propagated along the vein, indicating that cytosolic calcium ions act as long-distance rapid signals triggering leaf movements (see video). Furthermore, to investigate the evolutionary significance of these movements, they treated plants with a calcium channel blocker (La3+), which interfered with the touch response. These treated plants plants were more susceptible to herbivory by grasshoppers, demonstrating a defensive role for the leaf movements. (Summary by Mary Williams @PlantTeaching) Nature Comms. 10.1038/s41467-022-34106-x
Plants that respond to touch, like Mimosa pudica (aka the sensitive plant) are great ways to introduce children to the fascination of plants. If you haven’t had the chance to explore this plant, a touch to a leaflet causes the leaflets to fold into themselves, and recurring touches cause the leaf to drop away. Both responses occur due to a rapid loss of water from critical cells at hinge points. Hagihara et al. introduced into M. pudica an ultrasensitive fluorescent calcium ion (Ca2+) indicator (GCaMP6f) to investigate these fast responses. Wounding elicited a rapid increase in cytosolic calcium that propagated along the vein, indicating that cytosolic calcium ions act as long-distance rapid signals triggering leaf movements (see video). Furthermore, to investigate the evolutionary significance of these movements, they treated plants with a calcium channel blocker (La3+), which interfered with the touch response. These treated plants plants were more susceptible to herbivory by grasshoppers, demonstrating a defensive role for the leaf movements. (Summary by Mary Williams @PlantTeaching) Nature Comms. 10.1038/s41467-022-34106-x
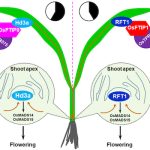
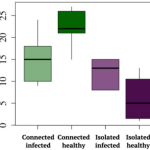
Staying connected helps plant population fitness
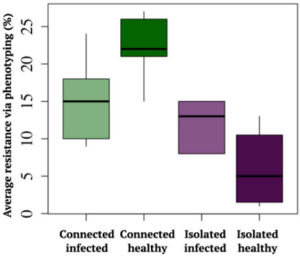 Many animals, including humans, tangibly benefit from living socially. Does this benefit extend to plants? A recent study by Höckerstedt et al. says it does. The authors studied a wild host-pathogen population, Plantago lanceolata, and its obligate powdery mildew pathogen Podosphaera plantaginis in 4000 locations in the Åland islands of Finland. They categorized the populations based on the previous infection and if the population was isolated or connected with other populations. From field studies, the authors observed that infection limits population growth in isolated populations more than in connected populations. Additionally, they set up an inoculation study using 19 isolated or well-connected populations and four different pathogen strains. From this, they observed that isolated populations are more susceptible and well-connected populations show a more diverse spectrum of resistance. The highest resistance was observed in previously uninfected connected populations while the least was observed in previously uninfected isolated populations. Transient and long-term simulations indicate that although disease prevalence, infectivity, and resistance will always be higher in well-connected populations, likely the evolutionary cost of maintaining the resistance will be lower. (Summary by Kamal Kumar Malukani, @KamalMalukani) Nature Comms. 10.1038/s41467-022-33665-3.
Many animals, including humans, tangibly benefit from living socially. Does this benefit extend to plants? A recent study by Höckerstedt et al. says it does. The authors studied a wild host-pathogen population, Plantago lanceolata, and its obligate powdery mildew pathogen Podosphaera plantaginis in 4000 locations in the Åland islands of Finland. They categorized the populations based on the previous infection and if the population was isolated or connected with other populations. From field studies, the authors observed that infection limits population growth in isolated populations more than in connected populations. Additionally, they set up an inoculation study using 19 isolated or well-connected populations and four different pathogen strains. From this, they observed that isolated populations are more susceptible and well-connected populations show a more diverse spectrum of resistance. The highest resistance was observed in previously uninfected connected populations while the least was observed in previously uninfected isolated populations. Transient and long-term simulations indicate that although disease prevalence, infectivity, and resistance will always be higher in well-connected populations, likely the evolutionary cost of maintaining the resistance will be lower. (Summary by Kamal Kumar Malukani, @KamalMalukani) Nature Comms. 10.1038/s41467-022-33665-3.