Plant Science Research Weekly: December 11, 2020
Review: The mechanical feedback theory of leaf lamina formation
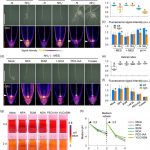
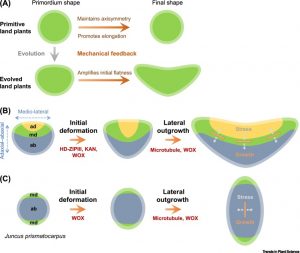 The contribution of microtubule orientation to the direction of cell expansion is familiar to most; when microtubules wrap around the middle of a cell like a belt, the cell expands in the perpendicular direction to become longer. Recent studies have extended this idea and proposed that the mechanical forces created by microtubules, along with reinforcing feedback, underly the development of flat leaves (see Zhao et al 2020). Here, Jiao et al. explore this model in the context of leaf evolution. They first consider Zimmermann’s telome theory, proposed more than 80 years ago, which attempted to describe how early plants with bifurcating radially symmetric branches (e.g., Rhynia), morphed towards megaphyll leaves. The authors observe that stages proposed by Zimmermann are difficult to correlate with our current understanding of leaf development. Rather, they suggest, mechanical feedback starting from a flattened primordium provides a better supported model for leaf blade evolution. (Summary by Mary Williams @PlantTeaching) Trends Plant Sci. 10.1016/j.tplants.2020.11.005
The contribution of microtubule orientation to the direction of cell expansion is familiar to most; when microtubules wrap around the middle of a cell like a belt, the cell expands in the perpendicular direction to become longer. Recent studies have extended this idea and proposed that the mechanical forces created by microtubules, along with reinforcing feedback, underly the development of flat leaves (see Zhao et al 2020). Here, Jiao et al. explore this model in the context of leaf evolution. They first consider Zimmermann’s telome theory, proposed more than 80 years ago, which attempted to describe how early plants with bifurcating radially symmetric branches (e.g., Rhynia), morphed towards megaphyll leaves. The authors observe that stages proposed by Zimmermann are difficult to correlate with our current understanding of leaf development. Rather, they suggest, mechanical feedback starting from a flattened primordium provides a better supported model for leaf blade evolution. (Summary by Mary Williams @PlantTeaching) Trends Plant Sci. 10.1016/j.tplants.2020.11.005
Review: Mesophyll conductance: walls, membranes and spatial complexity
 There’s been a lot of talk lately about how to improve the carboxylation efficiency of Rubisco, but of course this also depends on how much CO2 reaches the enzyme within the chloroplasts. To do so, it needs to pass through several distinct barriers: the boundary layer to reach the leaf surface, the stomatal pore, and then move within the leaf and into the mesophyll cell chloroplasts. In this Tansley Review, John Evans focusses on factors that affect this latter part, mesophyll conductance; these factors include cell walls and membranes. He addresses the molecular basis for differential mesophyll conductance, including wall composition and aquaporins, which are membrane channels that facilitate CO2 diffusion. He also addresses the feasibility of quantifying these factors through experimental and genetic studies as a basis to increase mesophyll conductance and to improve modeling towards the goal of improved photosynthesis. This is an exceptionally thorough and well-written review; instructors, consider adding it to your course reading list. (Summary by Mary Williams @PlantTeaching) New Phytol. 10.1111/nph.16968
There’s been a lot of talk lately about how to improve the carboxylation efficiency of Rubisco, but of course this also depends on how much CO2 reaches the enzyme within the chloroplasts. To do so, it needs to pass through several distinct barriers: the boundary layer to reach the leaf surface, the stomatal pore, and then move within the leaf and into the mesophyll cell chloroplasts. In this Tansley Review, John Evans focusses on factors that affect this latter part, mesophyll conductance; these factors include cell walls and membranes. He addresses the molecular basis for differential mesophyll conductance, including wall composition and aquaporins, which are membrane channels that facilitate CO2 diffusion. He also addresses the feasibility of quantifying these factors through experimental and genetic studies as a basis to increase mesophyll conductance and to improve modeling towards the goal of improved photosynthesis. This is an exceptionally thorough and well-written review; instructors, consider adding it to your course reading list. (Summary by Mary Williams @PlantTeaching) New Phytol. 10.1111/nph.16968
Combinations of maternal-specific repressive epigenetic marks in the endosperm control seed dormancy
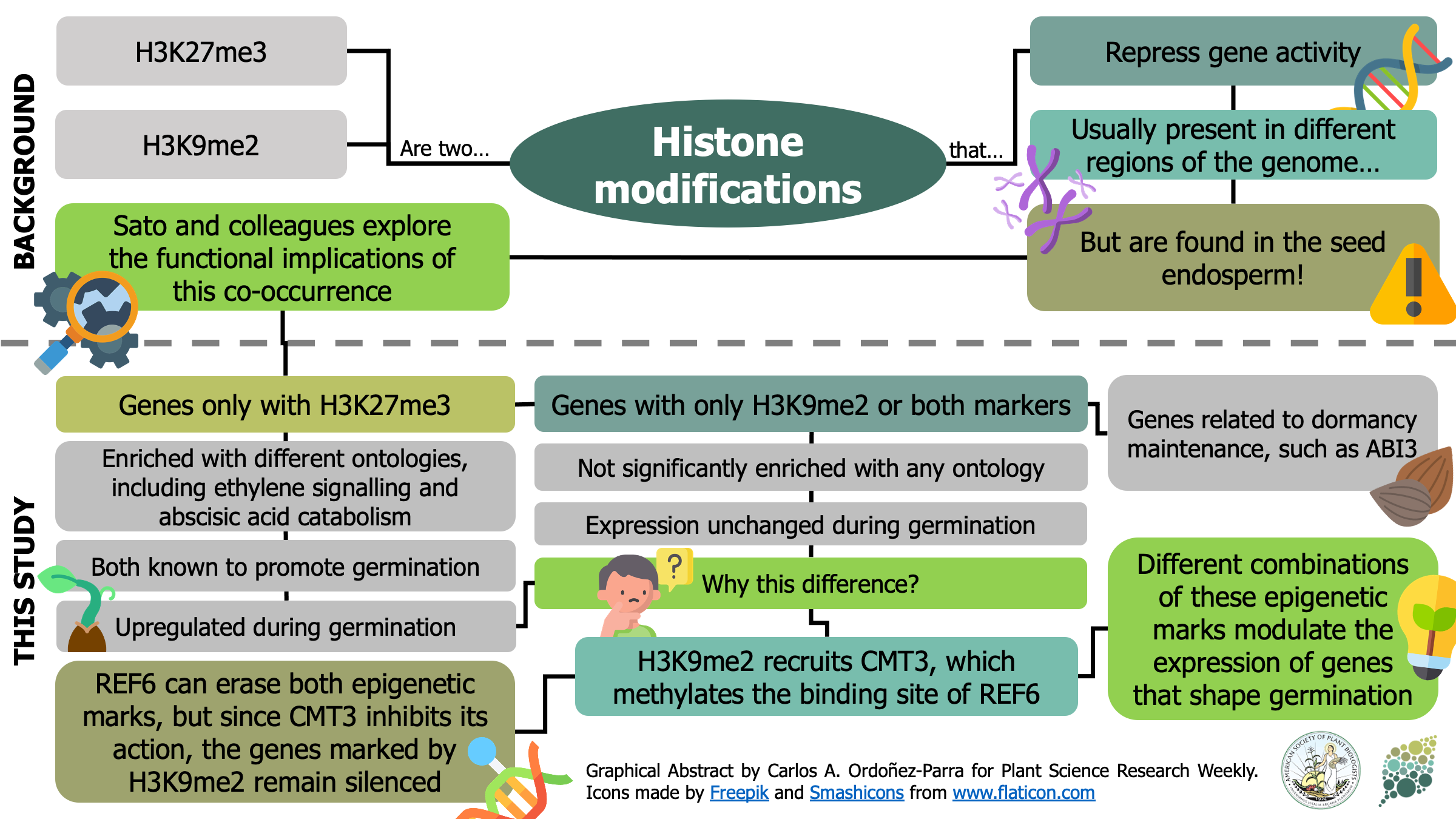 H3K27me3 [K27 (Lysine 27) trimethylation on histone H3] and H3K9me2 are two epigenetic modifications that repress gene activity in plants. While they are typically present in different genomic regions, both marks are found in the seed endosperm. Here, Sato and colleagues show that this histone modification duo is key to modulate gene activity during germination. Genes only marked with H3K27me3 were significantly enriched with gene ontologies including ethylene signaling and abscisic acid catabolism, both known for promoting germination. Moreover, they were found to be upregulated during germination process itself. Contrastingly, genes marked only with H3K9me2 or with both marks were not correlated to any ontology, nor had changes in their expression during germination. H3K9me2 recruits CHROMOMETHYL TRANSFERASE 3 (CMT3), which methylates the biding site of RELATIVE OF EARLY FLOWERING 6 (REF6) and inhibits its activity. Since REF6 can erase both H3K27me3 and H3K9me2 marks, CMT3 keeps marked genes inactive. Among the genes that present these two epigenetic marks, the authors found different dormancy maintenance genes such as ABA INSENSITIVE 3. Therefore, this research provides fascinating insights about how different combinations of epigenetic marks in the endosperm orchestrates the activation or silencing of genes that shape the germination process. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) bioRxiv. 10.1101/2020.11.10.376806
H3K27me3 [K27 (Lysine 27) trimethylation on histone H3] and H3K9me2 are two epigenetic modifications that repress gene activity in plants. While they are typically present in different genomic regions, both marks are found in the seed endosperm. Here, Sato and colleagues show that this histone modification duo is key to modulate gene activity during germination. Genes only marked with H3K27me3 were significantly enriched with gene ontologies including ethylene signaling and abscisic acid catabolism, both known for promoting germination. Moreover, they were found to be upregulated during germination process itself. Contrastingly, genes marked only with H3K9me2 or with both marks were not correlated to any ontology, nor had changes in their expression during germination. H3K9me2 recruits CHROMOMETHYL TRANSFERASE 3 (CMT3), which methylates the biding site of RELATIVE OF EARLY FLOWERING 6 (REF6) and inhibits its activity. Since REF6 can erase both H3K27me3 and H3K9me2 marks, CMT3 keeps marked genes inactive. Among the genes that present these two epigenetic marks, the authors found different dormancy maintenance genes such as ABA INSENSITIVE 3. Therefore, this research provides fascinating insights about how different combinations of epigenetic marks in the endosperm orchestrates the activation or silencing of genes that shape the germination process. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) bioRxiv. 10.1101/2020.11.10.376806
A novel GUN1-independent retrograde signaling pathway represses photomorphogenesis
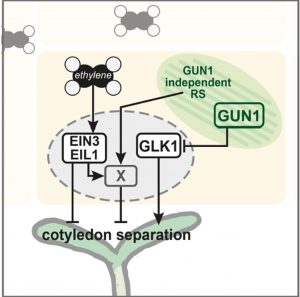 Seedlings emerging in light undergo photomorphogenic development, forming short hypocotyls and green, fully opened cotyledons. Disruption of chloroplast development by drugs such as lincomycin induces retrograde signals (RS) that inhibit photomorphogenesis. The retrograde signaling pathway has been known to act primarily through the chloroplast-localized master regulator protein GENOMES UNCOUPLED1 (GUN1). Now, Gommers et al. reveals the existence of a novel GUN1-independent pathway through which the retrograde signals induced by lincomycin inhibit cotyledon opening in light. This pathway relies on ethylene signaling that suppresses photomorphogenesis in seedlings. RS and ethylene signaling pathways converge at an unknown point downstream of the ethylene signaling transcription factors EIN3/EIL1 and co-induces a set of genes that are responsible for the inhibition of photomorphogenesis. These genes are distinct from the downstream genes of the light signaling network mediated by phytochromes and PIFs. The identity of the retrograde signal that is responsible for this novel pathway remains unknown. The authors speculate that it could be a known plastid-derived molecule like ROS, MecPP, PAP etc., or even a completely new signaling molecule that awaits discovery. (Summary by Yadukrishnan Premachandran @yadukrishprem) Plant Physiol. 10.1093/plphys/kiaa015
Seedlings emerging in light undergo photomorphogenic development, forming short hypocotyls and green, fully opened cotyledons. Disruption of chloroplast development by drugs such as lincomycin induces retrograde signals (RS) that inhibit photomorphogenesis. The retrograde signaling pathway has been known to act primarily through the chloroplast-localized master regulator protein GENOMES UNCOUPLED1 (GUN1). Now, Gommers et al. reveals the existence of a novel GUN1-independent pathway through which the retrograde signals induced by lincomycin inhibit cotyledon opening in light. This pathway relies on ethylene signaling that suppresses photomorphogenesis in seedlings. RS and ethylene signaling pathways converge at an unknown point downstream of the ethylene signaling transcription factors EIN3/EIL1 and co-induces a set of genes that are responsible for the inhibition of photomorphogenesis. These genes are distinct from the downstream genes of the light signaling network mediated by phytochromes and PIFs. The identity of the retrograde signal that is responsible for this novel pathway remains unknown. The authors speculate that it could be a known plastid-derived molecule like ROS, MecPP, PAP etc., or even a completely new signaling molecule that awaits discovery. (Summary by Yadukrishnan Premachandran @yadukrishprem) Plant Physiol. 10.1093/plphys/kiaa015
SAURs protein antagonistically regulate a phosphatase activity during apical hook development and cotyledon opening
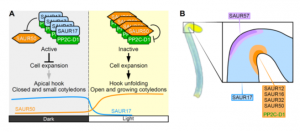 Upon germination in dark, plants adopt a strategy known as skotomorphogenesis, or etiolation, to protect the shoot apical meristem and cotyledon from damage while the seedling moves through the soil. The seedlings de-etiolate when they perceive light and the apical hook and cotyledon open. Wang et al. identified the Small Auxin Up RNA proteins SAUR17 and SAUR50 as cellular effectors of etiolation development. SAUR17 is highly expressed in the hook region and cotyledons of etiolated seedlings and serves to maintain the apical hook and closed cotyledons in dark. In contrast, SAUR50 promotes hook and cotyledon opening. Both SAURs bind to a clade D protein phosphatase 2C, PP2C-D1, a protein which also highly expressed in apical region and promotes cell expansion. Interestingly, SAUR17 binds more strongly to PP2C-D1 but does not inhibit its phosphatase activity, so functions as a protector to preserve PP2C-D1 activity against inhibition by SAUR50 in dark. Thus, the opposite functions of SAUR17 and SAUR50 to control apical hook development are mainly due to antagonistic regulation of PP2C-D1 in the dark. (Summary by Min May Wong @wongminmay) Plant Cell 10.1105/tpc.20.00283
Upon germination in dark, plants adopt a strategy known as skotomorphogenesis, or etiolation, to protect the shoot apical meristem and cotyledon from damage while the seedling moves through the soil. The seedlings de-etiolate when they perceive light and the apical hook and cotyledon open. Wang et al. identified the Small Auxin Up RNA proteins SAUR17 and SAUR50 as cellular effectors of etiolation development. SAUR17 is highly expressed in the hook region and cotyledons of etiolated seedlings and serves to maintain the apical hook and closed cotyledons in dark. In contrast, SAUR50 promotes hook and cotyledon opening. Both SAURs bind to a clade D protein phosphatase 2C, PP2C-D1, a protein which also highly expressed in apical region and promotes cell expansion. Interestingly, SAUR17 binds more strongly to PP2C-D1 but does not inhibit its phosphatase activity, so functions as a protector to preserve PP2C-D1 activity against inhibition by SAUR50 in dark. Thus, the opposite functions of SAUR17 and SAUR50 to control apical hook development are mainly due to antagonistic regulation of PP2C-D1 in the dark. (Summary by Min May Wong @wongminmay) Plant Cell 10.1105/tpc.20.00283
A grass-specific cellulose-xylan interaction dominates in sorghum secondary cell walls
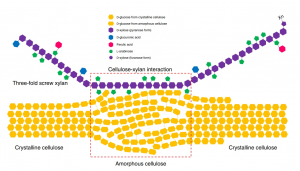 The cell walls of plants are intricate structures. Their complexity is due to multiple biochemical components and interactions, but while many of the components have been identified, their interactions have yet to be fully elucidated. A major component of the cell wall is cellulose, of which multiple subclasses exist. Xylan is the dominant hemicellulose, existing in two conformations: “two-fold screw” or “three-fold screw,” depending on their pattern of biochemical substitution, resulting in either a flat-ribbon shape or a helical shape, respectively. Previous reports utilized solid-state Nuclear Magnetic Resonance (ssNMR) techniques to demonstrate cell wall composition. In dicots, several studies demonstrated the presence of both two- and three-fold screw xylan, with two-fold xylan and crystalline cellulose interactions dominating the majority of xylan-cellulose interactions. However, the cell walls of monocot grasses such as sorghum are now of special interest, as they are huge renewable sources of feedstock for sustainable biorefineries, the energy and material producers of our future. Gao et al. performed ssNMR analysis on sorghum secondary cell walls. Interestingly, these cell walls present xylan primarily in the three-fold screw conformation. The authors conclude with a model highlighting three-screw xylan and amorphous cellulose interactions in the sorghum secondary cell wall, important information towards deciphering ideal methods for biomass deconstruction. (Summary by Benjamin Jin) Nature Communications 10.1038/s41467-020-19837-z
The cell walls of plants are intricate structures. Their complexity is due to multiple biochemical components and interactions, but while many of the components have been identified, their interactions have yet to be fully elucidated. A major component of the cell wall is cellulose, of which multiple subclasses exist. Xylan is the dominant hemicellulose, existing in two conformations: “two-fold screw” or “three-fold screw,” depending on their pattern of biochemical substitution, resulting in either a flat-ribbon shape or a helical shape, respectively. Previous reports utilized solid-state Nuclear Magnetic Resonance (ssNMR) techniques to demonstrate cell wall composition. In dicots, several studies demonstrated the presence of both two- and three-fold screw xylan, with two-fold xylan and crystalline cellulose interactions dominating the majority of xylan-cellulose interactions. However, the cell walls of monocot grasses such as sorghum are now of special interest, as they are huge renewable sources of feedstock for sustainable biorefineries, the energy and material producers of our future. Gao et al. performed ssNMR analysis on sorghum secondary cell walls. Interestingly, these cell walls present xylan primarily in the three-fold screw conformation. The authors conclude with a model highlighting three-screw xylan and amorphous cellulose interactions in the sorghum secondary cell wall, important information towards deciphering ideal methods for biomass deconstruction. (Summary by Benjamin Jin) Nature Communications 10.1038/s41467-020-19837-z
Allelic variation of MYB10 controls natural variation in skin and flesh color in strawberry
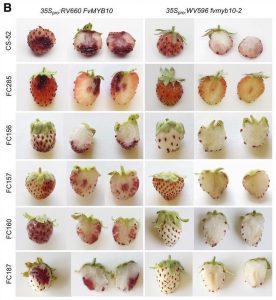 Few fruits have a more distinctive color than strawberry (Fragaria spp). Anthocyanins are responsible for strawberry’s characteristic red pigmentation with variations in receptacle color caused by altered anthocyanin levels. While the flavonoid synthesis pathway accountable for anthocyanin accumulation is well-characterized, a knowledge gap exists for the upstream network controlling this trait. Here, Castillejo et al. illustrate a simple system that controls natural variation in skin and flesh color in both diploid and octoploid strawberry. Quantitative trait loci mapping on an F2 population derived from a cross between white and red diploid accessions revealed that the MYB transcription factor MYB10 is the candidate gene for fruit color in strawberry. Subsequently, the authors investigated MYB10 allelic diversity in a geographically diverse ecotype panel. Three independent mutant alleles of MYB10 and a large chromosomal deletion were discovered that conferred the white fruit phenotype. A genome-wide association study in octoploid strawberry revealed anthocyanin biosynthesis to be activated primarily by an MYB10 copy- MYB10-2. A transposon insertion in the MYB10-2 promoter results in enhanced expression and thus anthocyanin accumulation in red fruit. MYB10 overexpression complemented the white fruit phenotypes of various accessions, making this gene a prime candidate for gene editing in strawberry. Finally, the authors developed markers to predict skin and flesh color that will likely facilitate molecular breeding in strawberry. (Summary by Caroline Dowling @CarolineD0wling) Plant Cell 10.1105/tpc.20.00474
Few fruits have a more distinctive color than strawberry (Fragaria spp). Anthocyanins are responsible for strawberry’s characteristic red pigmentation with variations in receptacle color caused by altered anthocyanin levels. While the flavonoid synthesis pathway accountable for anthocyanin accumulation is well-characterized, a knowledge gap exists for the upstream network controlling this trait. Here, Castillejo et al. illustrate a simple system that controls natural variation in skin and flesh color in both diploid and octoploid strawberry. Quantitative trait loci mapping on an F2 population derived from a cross between white and red diploid accessions revealed that the MYB transcription factor MYB10 is the candidate gene for fruit color in strawberry. Subsequently, the authors investigated MYB10 allelic diversity in a geographically diverse ecotype panel. Three independent mutant alleles of MYB10 and a large chromosomal deletion were discovered that conferred the white fruit phenotype. A genome-wide association study in octoploid strawberry revealed anthocyanin biosynthesis to be activated primarily by an MYB10 copy- MYB10-2. A transposon insertion in the MYB10-2 promoter results in enhanced expression and thus anthocyanin accumulation in red fruit. MYB10 overexpression complemented the white fruit phenotypes of various accessions, making this gene a prime candidate for gene editing in strawberry. Finally, the authors developed markers to predict skin and flesh color that will likely facilitate molecular breeding in strawberry. (Summary by Caroline Dowling @CarolineD0wling) Plant Cell 10.1105/tpc.20.00474
Spiral down: Rice plants adopt helical root growth under ammonium stress
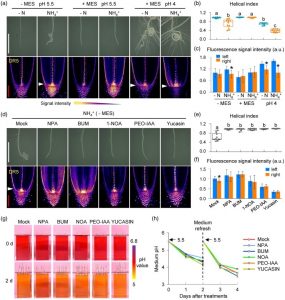 While ammonium ions (NH4+) serve as important sources of nitrogen nutrition, higher concentrations are toxic and inhibit plant growth and development. In an attempt to understand how roots of rice plant adapt to high concentrations of NH4+, Jia and colleagues found the roots coil and adopt a helical growth pattern at their tips. Experiments indicated this to be due to loss of gravitropism and it correlated with enhanced NH4+ uptake and the associated proton efflux and acidification of local medium. Detailed investigation of root auxin levels suggested that an asymmetric auxin distribution to one side of the root may result in the coiling pattern. Pharmacological studies too implicated an important role for auxin transport and signaling in promoting the helical growth of roots. Interestingly, Arabidopsis thaliana roots grown in medium with high NH4+ did not show the helical growth, suggesting it to be a particular adaptation in rice plants. (Summary by Pavithran Narayanan @pavi_narayanan) Plant J. 10.1111/tpj.14978
While ammonium ions (NH4+) serve as important sources of nitrogen nutrition, higher concentrations are toxic and inhibit plant growth and development. In an attempt to understand how roots of rice plant adapt to high concentrations of NH4+, Jia and colleagues found the roots coil and adopt a helical growth pattern at their tips. Experiments indicated this to be due to loss of gravitropism and it correlated with enhanced NH4+ uptake and the associated proton efflux and acidification of local medium. Detailed investigation of root auxin levels suggested that an asymmetric auxin distribution to one side of the root may result in the coiling pattern. Pharmacological studies too implicated an important role for auxin transport and signaling in promoting the helical growth of roots. Interestingly, Arabidopsis thaliana roots grown in medium with high NH4+ did not show the helical growth, suggesting it to be a particular adaptation in rice plants. (Summary by Pavithran Narayanan @pavi_narayanan) Plant J. 10.1111/tpj.14978