Plant Science Research Weekly: December 10, 2021
Review. Chloroplast development in green plant tissues: The interplay between light, hormones, and transcriptional regulation
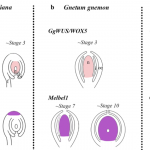
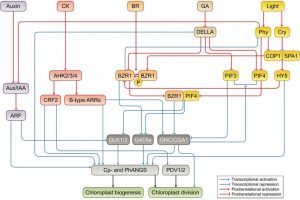 Chloroplasts are indispensable for plant growth and physiological performance; not only for photosynthesis but also for many biochemical processes. Due to the endosymbiont origin of the chloroplast, chloroplast development requires sophisticated machinery to relay the signals between the nuclear and the chloroplast genomes. Although the regulation of chloroplast biogenesis by light and phytohormones has been investigated, the molecular intricacies underlying integration of environmental and endogenous cues remain poorly understood. In this review, Cackett et al. highlight the key importance of transcription factors for interlinking multiple hormone and light signalling pathways to regulate chloroplast and photosynthesis-associated nuclear genes (Cp and PhANGs). The authors spotlight transcription factors such as GOLDEN2 LIKE-1 and 2 (GLK1&2), GATA NITRATE-INDUCIBLE CARBON-METABOLISM INVOLVED (GNC) and CYTOKININ-RESPONSIVE GATA FACTOR-1 (CGA-1) as the major regulatory nodes, while additional players need to be identified. Tuning these transcription regulatory nodes could be a rational approach to optimize chloroplast development, and thus photosynthetic efficiency. The authors conclude that further insights into the regulation of chloroplast development in tissue- and cell-specific manners awaits investigation. (Summary by Prakshi Aneja @PrakshiAneja) New Phytol. 10.1111/nph.17839
Chloroplasts are indispensable for plant growth and physiological performance; not only for photosynthesis but also for many biochemical processes. Due to the endosymbiont origin of the chloroplast, chloroplast development requires sophisticated machinery to relay the signals between the nuclear and the chloroplast genomes. Although the regulation of chloroplast biogenesis by light and phytohormones has been investigated, the molecular intricacies underlying integration of environmental and endogenous cues remain poorly understood. In this review, Cackett et al. highlight the key importance of transcription factors for interlinking multiple hormone and light signalling pathways to regulate chloroplast and photosynthesis-associated nuclear genes (Cp and PhANGs). The authors spotlight transcription factors such as GOLDEN2 LIKE-1 and 2 (GLK1&2), GATA NITRATE-INDUCIBLE CARBON-METABOLISM INVOLVED (GNC) and CYTOKININ-RESPONSIVE GATA FACTOR-1 (CGA-1) as the major regulatory nodes, while additional players need to be identified. Tuning these transcription regulatory nodes could be a rational approach to optimize chloroplast development, and thus photosynthetic efficiency. The authors conclude that further insights into the regulation of chloroplast development in tissue- and cell-specific manners awaits investigation. (Summary by Prakshi Aneja @PrakshiAneja) New Phytol. 10.1111/nph.17839
Extensive genome study to boost yield and improve agronomic traits in chickpea
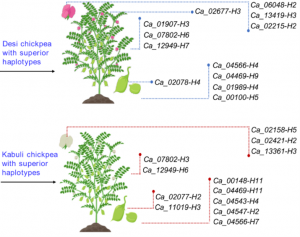 In recent years, there has been increasing awareness about the environmental impact of animal-based protein sources. Legumes such as chickpea (Cicer arientinum) are relatively cheap and sustainable sources of proteins, dietary fibres and micronutrients. Although there is a vast chickpea germplasm collection, little genomic characterization has been done. In Varshney et al., an international team of researchers from 41 organizations assembled the chickpea pan-genome by sequencing 3,366 accessions, includeing 3,171 cultivated and 195 wild varieties. A total of 29,870 genes, including 1582 novel genes, were identified in this study. Based on gene ontology analysis of the novel genes, several genes involved in adaptation to abiotic stresses have been identified. Further, the authors predicted the divergence and migration pattern of Cicer species across the globe based on the homology-based gene annotation. Most importantly, this study identified deleterious mutations and candidate genes responsible for reduced genetic diversity and fitness. The authors also report superior haplotypes (sets of DNA variations that tend not to recombine, and therefore are passed down through the generations together) in the local chickpea varieties that have distinctive characteristics arising from development and adaptation in a particular geographical area. This study provides a rich genetic resource for chickpea improvement and suggests ways to produce climate resilient and nutrient dense crops. (Summary by Chandan Kumar Gautam @chandan_gautam) Nature 10.1038/s41586-021-04066-1
In recent years, there has been increasing awareness about the environmental impact of animal-based protein sources. Legumes such as chickpea (Cicer arientinum) are relatively cheap and sustainable sources of proteins, dietary fibres and micronutrients. Although there is a vast chickpea germplasm collection, little genomic characterization has been done. In Varshney et al., an international team of researchers from 41 organizations assembled the chickpea pan-genome by sequencing 3,366 accessions, includeing 3,171 cultivated and 195 wild varieties. A total of 29,870 genes, including 1582 novel genes, were identified in this study. Based on gene ontology analysis of the novel genes, several genes involved in adaptation to abiotic stresses have been identified. Further, the authors predicted the divergence and migration pattern of Cicer species across the globe based on the homology-based gene annotation. Most importantly, this study identified deleterious mutations and candidate genes responsible for reduced genetic diversity and fitness. The authors also report superior haplotypes (sets of DNA variations that tend not to recombine, and therefore are passed down through the generations together) in the local chickpea varieties that have distinctive characteristics arising from development and adaptation in a particular geographical area. This study provides a rich genetic resource for chickpea improvement and suggests ways to produce climate resilient and nutrient dense crops. (Summary by Chandan Kumar Gautam @chandan_gautam) Nature 10.1038/s41586-021-04066-1
One assembly closer to understanding centromeric evolution in Arabidopsis thaliana
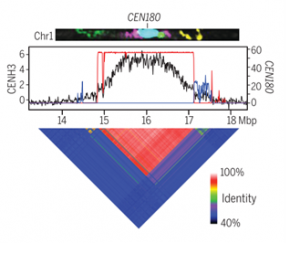 Centromeres are chromosome regions that consist of tandemly repeated “satellite arrays” and harbor CENH3 centromeric histones. Centromeres are present in every eukaryotic cell and are important for kinetochore assembly, so they ensure chromosome segregation during cell division. In spite of the importance of this structure, its evolution has remained obscure partly because of the challenge in sequencing these long and highly repeated sequences. Recently, this challenge has been overcome by new sequencing technologies. In this article, Naish and collaborators created a reference sequence of Arabidopsis thaliana centromeres, which are mainly composed of CEN180 satellite repeats. They successfully identified a library of 66,131 CEN180 repeat units and found that each chromosome presents a different arrangement of repeats and variants, suggesting different stages of adaptation and evolution. They also found that centromeres are invaded by young ATHILA retrotransposons, suggesting that retrotransposon insertion can influence centromere divergence. Finally, they found that the presence of CENH3 decreases non-CG methylation; similarly, methylation limits initiation of double strand breaks, the precursors of crossover events. These data suggest that the balance between the homogenization of CEN180 satellites and retrotransposon invasion drives centromeric evolution. (Summary by Rigel Salinas-Gamboa @Rigelitactica) Science 10.1126/science.abi7489
Centromeres are chromosome regions that consist of tandemly repeated “satellite arrays” and harbor CENH3 centromeric histones. Centromeres are present in every eukaryotic cell and are important for kinetochore assembly, so they ensure chromosome segregation during cell division. In spite of the importance of this structure, its evolution has remained obscure partly because of the challenge in sequencing these long and highly repeated sequences. Recently, this challenge has been overcome by new sequencing technologies. In this article, Naish and collaborators created a reference sequence of Arabidopsis thaliana centromeres, which are mainly composed of CEN180 satellite repeats. They successfully identified a library of 66,131 CEN180 repeat units and found that each chromosome presents a different arrangement of repeats and variants, suggesting different stages of adaptation and evolution. They also found that centromeres are invaded by young ATHILA retrotransposons, suggesting that retrotransposon insertion can influence centromere divergence. Finally, they found that the presence of CENH3 decreases non-CG methylation; similarly, methylation limits initiation of double strand breaks, the precursors of crossover events. These data suggest that the balance between the homogenization of CEN180 satellites and retrotransposon invasion drives centromeric evolution. (Summary by Rigel Salinas-Gamboa @Rigelitactica) Science 10.1126/science.abi7489
Prediction of conserved variable heat and cold stress response in maize using cis-regulatory information
 Several transcription factors (TFs) have been identified previously as responsive to cold and heat stresses in maize. Variation in TF binding sites (TFBS) determine changes in the expression of target genes in response to the aforementioned stresses. Traditionally, single varieties were used to study TFBS variation rather than different species which are, putatively, a source of cis-regulatory variation. To deepen this aspect, in this work Zhou et al. compared inbred and hybrid genotype responses after applying different heat/cold stresses. In addition, they developed machine learning models for predicting gene expression responses in new genotypes, highlighting the critical parameters for motif detection. To conclude, they point out that insertion/deletion (InDel) polymorphisms have a fundamental role in the cis-regulatory elements of the genes with differential responses to heat/cold stress. This work is a step further to study the link between the cis-regulatory variation, gene expression, and plant phenotypes. (Summary by Eva Maria Gomez Alvarez, @eva_ga96) Plant Cell 10.1093/plcell/koab267.
Several transcription factors (TFs) have been identified previously as responsive to cold and heat stresses in maize. Variation in TF binding sites (TFBS) determine changes in the expression of target genes in response to the aforementioned stresses. Traditionally, single varieties were used to study TFBS variation rather than different species which are, putatively, a source of cis-regulatory variation. To deepen this aspect, in this work Zhou et al. compared inbred and hybrid genotype responses after applying different heat/cold stresses. In addition, they developed machine learning models for predicting gene expression responses in new genotypes, highlighting the critical parameters for motif detection. To conclude, they point out that insertion/deletion (InDel) polymorphisms have a fundamental role in the cis-regulatory elements of the genes with differential responses to heat/cold stress. This work is a step further to study the link between the cis-regulatory variation, gene expression, and plant phenotypes. (Summary by Eva Maria Gomez Alvarez, @eva_ga96) Plant Cell 10.1093/plcell/koab267.
The main oxidative inactivation pathway of the plant hormone auxin
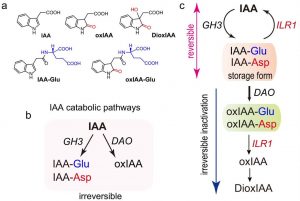 Auxin is a key hormone in almost all stages of the plant development and its main natural form is indole-3-acetic acid (IAA). In Arabidopsis, several gene families are involved in auxin homeostasis. In this work, Hayashi et al. focus on the roles of DIOXYGENASE FOR AUXIN OXIDATION 1 (DAO1) which converts IAA to oxIAA; the GH3 gene family encoding for acyl amidotransferases that catalyze the conjugation of IAA with amino acids (forming IAA-Asp and IAA-Glu); and IAA-Leu-Resistant1 (ILR1), a Leu-amidohydrolase which converts IAA-amino acid conjugates to IAA in vitro. Here, the authors describe how IAA is inactivated and recycled through the GH3-ILR1-DAO pathway. Basically, GH3 converts IAA to the storage form IAA-Asp and IAA-Glu, a process that can be reversed by ILR1. If needed, IAA-Asp and IAA-Glu can be oxidized by DAO1 in an irreversible reaction. Then, ILR1 catalyzes the release of the amino acids from oxIAA. This work describes an auxin inactivation pathway that might be important for auxin homeostasis. (Summary by Eva Maria Gomez Alvarez, @eva_ga96) Nature Comms. https://doi.org/10.1038/s41467-021-27020-1.
Auxin is a key hormone in almost all stages of the plant development and its main natural form is indole-3-acetic acid (IAA). In Arabidopsis, several gene families are involved in auxin homeostasis. In this work, Hayashi et al. focus on the roles of DIOXYGENASE FOR AUXIN OXIDATION 1 (DAO1) which converts IAA to oxIAA; the GH3 gene family encoding for acyl amidotransferases that catalyze the conjugation of IAA with amino acids (forming IAA-Asp and IAA-Glu); and IAA-Leu-Resistant1 (ILR1), a Leu-amidohydrolase which converts IAA-amino acid conjugates to IAA in vitro. Here, the authors describe how IAA is inactivated and recycled through the GH3-ILR1-DAO pathway. Basically, GH3 converts IAA to the storage form IAA-Asp and IAA-Glu, a process that can be reversed by ILR1. If needed, IAA-Asp and IAA-Glu can be oxidized by DAO1 in an irreversible reaction. Then, ILR1 catalyzes the release of the amino acids from oxIAA. This work describes an auxin inactivation pathway that might be important for auxin homeostasis. (Summary by Eva Maria Gomez Alvarez, @eva_ga96) Nature Comms. https://doi.org/10.1038/s41467-021-27020-1.
The calcium-dependent protein kinase CPK28 is targeted by the ubiquitin ligases ATL31 and ATL6 for proteasome-mediated degradation to fine-tune immune signaling in Arabidopsis
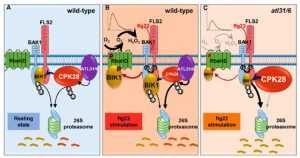 ‘Fine tuning’ is a very significant process that takes place in the cell to correctly modulate plant responses. BOTRYTIS-INDUCED KINASE 1 (BIK1) is an important hub protein that is at the center of the signaling hub that controls defense responses. Hence, plants fine tune this important protein through several processes, including proteasomal degradation mediated by a Ca2+ dependent protein kinase CDPK28 during the absence of stress. In a recent report by Liu et al. in The Plant Cell, the authors show that the CPK28 protein is itself regulated by proteasomal degradation to free up BIK1 to elicit the defense response. Under pathogen attack, which the authors elicited by flagellin 22 (flg22) treatment, two ubiquitin ligases ATL31 and ATL6 interact with CPK28 and to promote its degradation through the 26S proteasome system. This frees up significant amount of BIK1 protein that can now activate RbohD by phosphorylation, leading to ROS production. In separate events, BIK1 can also activate other processes, including the closure of stomata and the initation of the Ca2+ signaling cascade, to further enhance the plant’s defense response. (Summary by Sibaji K Sanyal) Plant Cell 10.1093/plcell/koab242
‘Fine tuning’ is a very significant process that takes place in the cell to correctly modulate plant responses. BOTRYTIS-INDUCED KINASE 1 (BIK1) is an important hub protein that is at the center of the signaling hub that controls defense responses. Hence, plants fine tune this important protein through several processes, including proteasomal degradation mediated by a Ca2+ dependent protein kinase CDPK28 during the absence of stress. In a recent report by Liu et al. in The Plant Cell, the authors show that the CPK28 protein is itself regulated by proteasomal degradation to free up BIK1 to elicit the defense response. Under pathogen attack, which the authors elicited by flagellin 22 (flg22) treatment, two ubiquitin ligases ATL31 and ATL6 interact with CPK28 and to promote its degradation through the 26S proteasome system. This frees up significant amount of BIK1 protein that can now activate RbohD by phosphorylation, leading to ROS production. In separate events, BIK1 can also activate other processes, including the closure of stomata and the initation of the Ca2+ signaling cascade, to further enhance the plant’s defense response. (Summary by Sibaji K Sanyal) Plant Cell 10.1093/plcell/koab242
Expression analyses in Ginkgo biloba provide new insights into the evolution and development of the seed
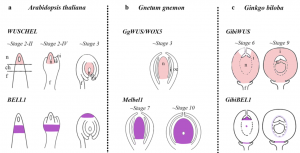 Seed production was a decisive adaptation that appeared during plant evolution. Not in vain! The most diverse plant lineage today is that of the so-called seed plants, a group that includes gymnosperms and angiosperms. Therefore, it is not surprising that understanding seed origin and evolution has been of paramount interest within the plant science community for a long time. However, most of the available information about the molecular mechanisms behind seed development is limited to angiosperms. In this fascinating research, Zumajo-Cardona and colleagues conducted a thorough expression analysis during the seed development of Gingko biloba –a “living-fossil” gymnosperm present in one of the first lineages where seeds evolved. A spatiotemporal analysis of six genes known for their functions in angiosperm seed development revealed that expression patterns are not fully conserved between angiosperms and gymnosperms. Notably, several genes restricted to ovules integuments in angiosperms were expressed in developing sporangia in Ginkgo and non-seed plant lineages, implying that the integument developmental network might have been co-opted from the ones controlling sporangia development. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Sci. Rep. 10.1038/s41598-021-01483-0.
Seed production was a decisive adaptation that appeared during plant evolution. Not in vain! The most diverse plant lineage today is that of the so-called seed plants, a group that includes gymnosperms and angiosperms. Therefore, it is not surprising that understanding seed origin and evolution has been of paramount interest within the plant science community for a long time. However, most of the available information about the molecular mechanisms behind seed development is limited to angiosperms. In this fascinating research, Zumajo-Cardona and colleagues conducted a thorough expression analysis during the seed development of Gingko biloba –a “living-fossil” gymnosperm present in one of the first lineages where seeds evolved. A spatiotemporal analysis of six genes known for their functions in angiosperm seed development revealed that expression patterns are not fully conserved between angiosperms and gymnosperms. Notably, several genes restricted to ovules integuments in angiosperms were expressed in developing sporangia in Ginkgo and non-seed plant lineages, implying that the integument developmental network might have been co-opted from the ones controlling sporangia development. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Sci. Rep. 10.1038/s41598-021-01483-0.