Plant Science Research Weekly: August 30th
A modular cloning toolkit for genome editing in plants
 Genome editing with the CRISPR/Cas system is now widely used in functional studies across biological sciences including plant biology. Typically, this system involves a DNA nuclease and a guide RNA that directs the nuclease to a specific location in the genome. Golden Gate (GG) is a cloning method that is able to efficiently assemble multiple expression units and has been used successfully in plant genome editing applications. However, GG modules for some of the latest genome editing enzymes are lacking. Here, Hahn et al. have developed an additional cloning toolkit which contains ninety-nine modules encoding several CRISPR/Cas nucleases and their cognate guide RNA. In addition, the authors developed other genetic components including several promoters for driving the expression of CRISPR/Cas nuclease in monocots and dicots. Lastly, they also developed a set of modules that allows the construction of tRNA-sgRNA polycistronic units and demonstrated the application of this system in wheat. The resources are available from Addgene. (Summary by Toluwase Olukayode) bioRxiv 10.1101/738021
Genome editing with the CRISPR/Cas system is now widely used in functional studies across biological sciences including plant biology. Typically, this system involves a DNA nuclease and a guide RNA that directs the nuclease to a specific location in the genome. Golden Gate (GG) is a cloning method that is able to efficiently assemble multiple expression units and has been used successfully in plant genome editing applications. However, GG modules for some of the latest genome editing enzymes are lacking. Here, Hahn et al. have developed an additional cloning toolkit which contains ninety-nine modules encoding several CRISPR/Cas nucleases and their cognate guide RNA. In addition, the authors developed other genetic components including several promoters for driving the expression of CRISPR/Cas nuclease in monocots and dicots. Lastly, they also developed a set of modules that allows the construction of tRNA-sgRNA polycistronic units and demonstrated the application of this system in wheat. The resources are available from Addgene. (Summary by Toluwase Olukayode) bioRxiv 10.1101/738021
The N-terminus of AtMSL10 interacts with its own C- terminus
 Plants are equipped with multiple mechanosensitive (MS) ion channels that respond to external and internal mechanical perturbations. When one of these, AtMSL10, is overexpressed it leads to a cell death phenotype, although there is no discernible phenotype associated with its loss of function. Recently Basu et al. demonstrated a novel activation mechanism of AtMSL10 when transducing the effect of increased membrane tension: when the ion channel opened in response to membrane tension, a conformational change at MSL10 C-terminus occurred. This structural rearrangement causes the dephosphorylating of the N-terminus of AtMSL10, activated it and triggering MSL10-directed cell death. The authors also showed that AtMSL10 activation induces expression of several stress-responsive genes. This study is interesting for proposing a mechanism involving the intra- and/intermolecular interaction between N- and C- termini of the same protein and it also sheds lights on the process of how plants perceive and respond to stress. (Summary by Nanxun Qin) bioRxiv 10.1101/726521
Plants are equipped with multiple mechanosensitive (MS) ion channels that respond to external and internal mechanical perturbations. When one of these, AtMSL10, is overexpressed it leads to a cell death phenotype, although there is no discernible phenotype associated with its loss of function. Recently Basu et al. demonstrated a novel activation mechanism of AtMSL10 when transducing the effect of increased membrane tension: when the ion channel opened in response to membrane tension, a conformational change at MSL10 C-terminus occurred. This structural rearrangement causes the dephosphorylating of the N-terminus of AtMSL10, activated it and triggering MSL10-directed cell death. The authors also showed that AtMSL10 activation induces expression of several stress-responsive genes. This study is interesting for proposing a mechanism involving the intra- and/intermolecular interaction between N- and C- termini of the same protein and it also sheds lights on the process of how plants perceive and respond to stress. (Summary by Nanxun Qin) bioRxiv 10.1101/726521
Epigenetic silencing of a multifunctional plant stress regulator EIN2
 EIN2 is a key regulator of the ethylene response and a protein with a complex function and regulation. Zander et al. have uncovered an interesting and complex mechanism by which its expression is regulated. Many genes are regulated through histone modifications, with the histone variant H2A.Z and the histone mark H3K27me3 both repressors of transcription. The ein6-1 mutant is a fast-neutron generated mutant that the authors have now shown carries mutations in two different unlinked genes. The ethylene hyposensitive phenotype occurs in what the authors refer to as the ein6-1 single mutant; this gene encodes a histone methyltransferase. The second mutation is in a gene they have named EIN6 ENHANCER (EEN), which alone has no phenotype but which enhances the ein6-1 phenotype. They show that in wild-type plants, these two gene products work cooperatively to remove repressive chromatin marks, and when both are mutated the EIN2 gene is not expressed. They conclude, “These results uncover a unique type of chromatin regulation which safeguards the expression of an essential multifunctional plant stress regulator.” (Summary by Mary Williams) eLIFE 10.7554/eLife.47835
EIN2 is a key regulator of the ethylene response and a protein with a complex function and regulation. Zander et al. have uncovered an interesting and complex mechanism by which its expression is regulated. Many genes are regulated through histone modifications, with the histone variant H2A.Z and the histone mark H3K27me3 both repressors of transcription. The ein6-1 mutant is a fast-neutron generated mutant that the authors have now shown carries mutations in two different unlinked genes. The ethylene hyposensitive phenotype occurs in what the authors refer to as the ein6-1 single mutant; this gene encodes a histone methyltransferase. The second mutation is in a gene they have named EIN6 ENHANCER (EEN), which alone has no phenotype but which enhances the ein6-1 phenotype. They show that in wild-type plants, these two gene products work cooperatively to remove repressive chromatin marks, and when both are mutated the EIN2 gene is not expressed. They conclude, “These results uncover a unique type of chromatin regulation which safeguards the expression of an essential multifunctional plant stress regulator.” (Summary by Mary Williams) eLIFE 10.7554/eLife.47835
Coordination of circadian times in Arabidopsis seedlings
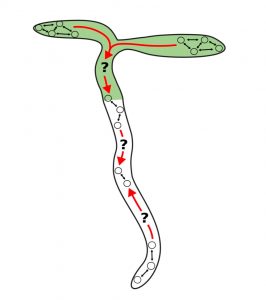 Precise temporal control is crucial for plant developmental processes, e.g., photosynthesis, leaf movement, and flowering. In this work, Greenwood et al. studied how rhythms are coordinated at the whole-organism level. The authors monitored the activity of GIGANTEA (GI), a core clock gene, using a LUCIFERASE reporter gene in Arabidopsis seedlings. GI:LUC expression showed different peaks across the plant, indicating different clock rhythms among organs. Several spatial waves of GI gene expression were detected in the plant. In the cotyledons the wave propagated from the tip down through the hypocotyl, and another wave started from the root junction going upwards. Additionally, two different waves were detected in the roots, one starting in the hypocotyl junction going downwards and another from the root tip moving upwards. To test if these expression waves were driven by long-distance communication, the authors monitored the rhythms in excised cotyledons, roots, and root tips, and found that clocks across the plant are coordinated via local cell-cell signaling and are independent of a long-distance signal. Finally, they tested different metabolic inputs (e.g., sugars, inhibitors of photosynthesis) and genetic elements (e.g., phytochrome mutants) to investigate their effects on plant-wide circadian clock gene expression. (Summary by Humberto Herrera-Ubaldo) PLOS Biol. 10.1371/journal.pbio.3000407
Precise temporal control is crucial for plant developmental processes, e.g., photosynthesis, leaf movement, and flowering. In this work, Greenwood et al. studied how rhythms are coordinated at the whole-organism level. The authors monitored the activity of GIGANTEA (GI), a core clock gene, using a LUCIFERASE reporter gene in Arabidopsis seedlings. GI:LUC expression showed different peaks across the plant, indicating different clock rhythms among organs. Several spatial waves of GI gene expression were detected in the plant. In the cotyledons the wave propagated from the tip down through the hypocotyl, and another wave started from the root junction going upwards. Additionally, two different waves were detected in the roots, one starting in the hypocotyl junction going downwards and another from the root tip moving upwards. To test if these expression waves were driven by long-distance communication, the authors monitored the rhythms in excised cotyledons, roots, and root tips, and found that clocks across the plant are coordinated via local cell-cell signaling and are independent of a long-distance signal. Finally, they tested different metabolic inputs (e.g., sugars, inhibitors of photosynthesis) and genetic elements (e.g., phytochrome mutants) to investigate their effects on plant-wide circadian clock gene expression. (Summary by Humberto Herrera-Ubaldo) PLOS Biol. 10.1371/journal.pbio.3000407
Jasmonate-related MYC transcription factors are functionally conserved in Marchantia polymorpha ($)
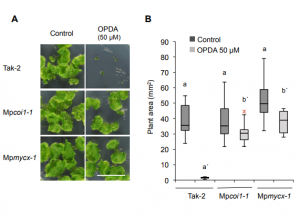 In recent years, studies in the model bryophyte Marchantia polymorpha have provided insights on how the Jasmonic Acid (JA) pathway works in early diverging land plants, starting from its biosynthesis (Yamamoto et al., 2015; Koeduka et al., 2015), followed by perception (Monte et al, 2018) and co-repressor (Monte et al, 2019), and now, the transcription factors involved in the downstream signaling. Marchantia possesses two homologs of AtMYC2, the master regulator in Arabidopsis: MpMYCX and MpMYCY, which are encoded in the X and Y chromosomes and expressed in the male and female gametophyte, respectively. Here, Peñuelas et al. show that the role of these homologs is conserved in Marchantia, including interactions with JAZ proteins, light stability, and phenotypic effects on dino-OPDA (the functional JA in Marchantia). The disruption of JA signaling pathways makes Marchantia plants more sensitive to herbivory, and mutants in Mpmycy accumulate a reduced amount of sesquiterpenoids in response to herbivory, which suggests that terpenoid biosynthesis could be a defense mechanism conserved also in Marchantia. Additionally, the authors found that there is no reciprocal inter-specific complementation of MYCs between Arabidopsis and Marchantia. This finding shows how hormone pathways could be conserved across embryophytes, but some protein-protein interactions would have changed along with evolution. (Summary by Facundo Romani) Plant Cell 10.1105/tpc.18.00974. See also In Brief by Phil Carella 10.1105/tpc.19.00600.
In recent years, studies in the model bryophyte Marchantia polymorpha have provided insights on how the Jasmonic Acid (JA) pathway works in early diverging land plants, starting from its biosynthesis (Yamamoto et al., 2015; Koeduka et al., 2015), followed by perception (Monte et al, 2018) and co-repressor (Monte et al, 2019), and now, the transcription factors involved in the downstream signaling. Marchantia possesses two homologs of AtMYC2, the master regulator in Arabidopsis: MpMYCX and MpMYCY, which are encoded in the X and Y chromosomes and expressed in the male and female gametophyte, respectively. Here, Peñuelas et al. show that the role of these homologs is conserved in Marchantia, including interactions with JAZ proteins, light stability, and phenotypic effects on dino-OPDA (the functional JA in Marchantia). The disruption of JA signaling pathways makes Marchantia plants more sensitive to herbivory, and mutants in Mpmycy accumulate a reduced amount of sesquiterpenoids in response to herbivory, which suggests that terpenoid biosynthesis could be a defense mechanism conserved also in Marchantia. Additionally, the authors found that there is no reciprocal inter-specific complementation of MYCs between Arabidopsis and Marchantia. This finding shows how hormone pathways could be conserved across embryophytes, but some protein-protein interactions would have changed along with evolution. (Summary by Facundo Romani) Plant Cell 10.1105/tpc.18.00974. See also In Brief by Phil Carella 10.1105/tpc.19.00600.
A peptide pair coordinates ovule initiation patterns with seed number and fruit size
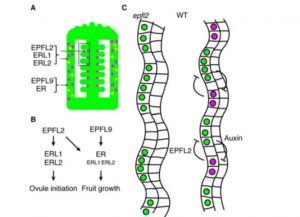 Plant peptide patterning is popping up everywhere! Now Kawamoto et al. show that a pair of peptides, interacting with receptor proteins, coordinates the placement of ovules in Arabidopsis. They started with a natural variation analysis, which highlighted that several varieties with mutations in the ERECTA gene (which encodes a LRR-receptor kinase) have high-ovule densities, which is revertible upon introduction of the wild-type ER gene. ERECTA and related LRR-receptors are known to bind peptide ligands. The authors found one, EPFL9 (also known as STOMAGEN) that is expressed in developing fruit and when knocked-down by RNAi leads to an increase in ovule density. The authors also identify a module involving ERECTA-Like proteins and another peptide as important in ovule patterning, and propose a model involving patterning through negative feedback with auxin. Finally, they note that a related peptide has been shown to control rice grain number and size, suggesting a universality of this mechanism. (Summary by Mary Williams) bioRxiv 10.1101/736439
Plant peptide patterning is popping up everywhere! Now Kawamoto et al. show that a pair of peptides, interacting with receptor proteins, coordinates the placement of ovules in Arabidopsis. They started with a natural variation analysis, which highlighted that several varieties with mutations in the ERECTA gene (which encodes a LRR-receptor kinase) have high-ovule densities, which is revertible upon introduction of the wild-type ER gene. ERECTA and related LRR-receptors are known to bind peptide ligands. The authors found one, EPFL9 (also known as STOMAGEN) that is expressed in developing fruit and when knocked-down by RNAi leads to an increase in ovule density. The authors also identify a module involving ERECTA-Like proteins and another peptide as important in ovule patterning, and propose a model involving patterning through negative feedback with auxin. Finally, they note that a related peptide has been shown to control rice grain number and size, suggesting a universality of this mechanism. (Summary by Mary Williams) bioRxiv 10.1101/736439
The ectomycorrhizal fungus Laccaria bicolor produces lipochitooligosaccharides and uses the common symbiosis pathway ($)
 Nutrient exchange during plant-fungal symbiosis allows assimilation of nutrients necessary for plant growth in a beneficial way. Several plants have evolved to form symbioses with mycorrhizal fungi and rhizobia bacteria that allow the plants to efficiently take up nitrogen and phosphorous. In its interactions with mycorrhizal fungi, the plant secretes strigolactones (hormones) that are perceived by the fungus, which in return secretes lipochitooligosaccharides (LCOs). Exchange of these signals triggers a signaling cascade in plants that allows symbiosis to succeed. Previous studies have identified a common symbiosis pathway that is required for the interaction between plants and arbuscular mycorrhizal fungi or rhizobia bacteria. Cope et al. have demonstrated the conservation of the common symbiosis pathway in the interaction between the model woody plant Populus and the ectomycorrhizal fungus Laccaria bicolor. Using a genetic approach they showed the involvement of key conserved plant genes. Using mass-spectrometry approaches, the authors have identified the potential LCOs secreted by the fungal partner. Host phenotypic changes, such as root hair response and molecular changes occurring during symbiosis, demonstrate that a plant-ectomycorrhizal relationship has been established. (Summary by Suresh Damodaran) Plant Cell 10.1105/tpc.18.00676
Nutrient exchange during plant-fungal symbiosis allows assimilation of nutrients necessary for plant growth in a beneficial way. Several plants have evolved to form symbioses with mycorrhizal fungi and rhizobia bacteria that allow the plants to efficiently take up nitrogen and phosphorous. In its interactions with mycorrhizal fungi, the plant secretes strigolactones (hormones) that are perceived by the fungus, which in return secretes lipochitooligosaccharides (LCOs). Exchange of these signals triggers a signaling cascade in plants that allows symbiosis to succeed. Previous studies have identified a common symbiosis pathway that is required for the interaction between plants and arbuscular mycorrhizal fungi or rhizobia bacteria. Cope et al. have demonstrated the conservation of the common symbiosis pathway in the interaction between the model woody plant Populus and the ectomycorrhizal fungus Laccaria bicolor. Using a genetic approach they showed the involvement of key conserved plant genes. Using mass-spectrometry approaches, the authors have identified the potential LCOs secreted by the fungal partner. Host phenotypic changes, such as root hair response and molecular changes occurring during symbiosis, demonstrate that a plant-ectomycorrhizal relationship has been established. (Summary by Suresh Damodaran) Plant Cell 10.1105/tpc.18.00676
Increased atmospheric vapor pressure deficit reduces global vegetation growth
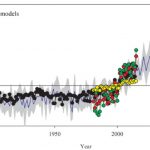
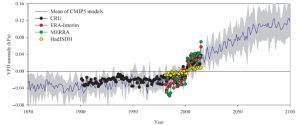 Plant scientists know that when the air at the leaf surface is dry, the plant will tend to close its stomata, but we often consider this in terms of its short-term, localized effects. Yuan et al. have explored the global trends in vapor pressure deficit (VPD; difference between saturated and real water content of the air) and found that the VPD of vegetated areas has been rising notably since the late 1990s. This anomaly is due to both increased temperatures (which increase saturated water vapor pressure) and decreased actual water vapor pressure. As water vapor over land is mostly derived from oceanic evaporation, they also looked at this trend and found a transition from upward to downward also in the late 1990s. The authors observe that the increase in VPD is likely to have contributed to forest mortality (in part due to reduced carbon assimilation, as well as drought effects) in the past decades, and also that most current models fail to include the effect of VPD, meaning they are likely to overestimate future plant growth and carbon sequestration. (Summary by Mary Williams) Science Advances 10.1126/sciadv.aax1396
Plant scientists know that when the air at the leaf surface is dry, the plant will tend to close its stomata, but we often consider this in terms of its short-term, localized effects. Yuan et al. have explored the global trends in vapor pressure deficit (VPD; difference between saturated and real water content of the air) and found that the VPD of vegetated areas has been rising notably since the late 1990s. This anomaly is due to both increased temperatures (which increase saturated water vapor pressure) and decreased actual water vapor pressure. As water vapor over land is mostly derived from oceanic evaporation, they also looked at this trend and found a transition from upward to downward also in the late 1990s. The authors observe that the increase in VPD is likely to have contributed to forest mortality (in part due to reduced carbon assimilation, as well as drought effects) in the past decades, and also that most current models fail to include the effect of VPD, meaning they are likely to overestimate future plant growth and carbon sequestration. (Summary by Mary Williams) Science Advances 10.1126/sciadv.aax1396
Large-effect flowering time mutations reveal conditionally adaptive paths through fitness landscapes in Arabidopsis thaliana
 There is a tendency to think of genes carrying mutations as having a negative impact on fitness, which raises the question of why they might persist in a population. Taylor et al. tested whether large-effect mutations that affect flowering time might not be detrimental in all conditions, by comparing their effects in natural populations of Arabidopsis grown in different environments. They selected several genes that have large effects on flowering time, including FT, light signaling, including PHYA, and branching, including SPY, and they grew them alongside other genotypes in several outdoor field sites across, and then measured bolting time, branching and fecundity. They found a great deal of plasticity in how these genes affected the measured traits, and that the relative fitness amongst mutants varied with the different environments. They conclude, “large effect mutations may persist in populations because they influence traits that are adaptive only under specific environmental conditions.” (Summary by Mary Williams) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1902731116
There is a tendency to think of genes carrying mutations as having a negative impact on fitness, which raises the question of why they might persist in a population. Taylor et al. tested whether large-effect mutations that affect flowering time might not be detrimental in all conditions, by comparing their effects in natural populations of Arabidopsis grown in different environments. They selected several genes that have large effects on flowering time, including FT, light signaling, including PHYA, and branching, including SPY, and they grew them alongside other genotypes in several outdoor field sites across, and then measured bolting time, branching and fecundity. They found a great deal of plasticity in how these genes affected the measured traits, and that the relative fitness amongst mutants varied with the different environments. They conclude, “large effect mutations may persist in populations because they influence traits that are adaptive only under specific environmental conditions.” (Summary by Mary Williams) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1902731116