Plant Science Research Weekly: August 27, 2021
Review: Alternative splicing as conserved mechanism to regulate CDPKs? ($)
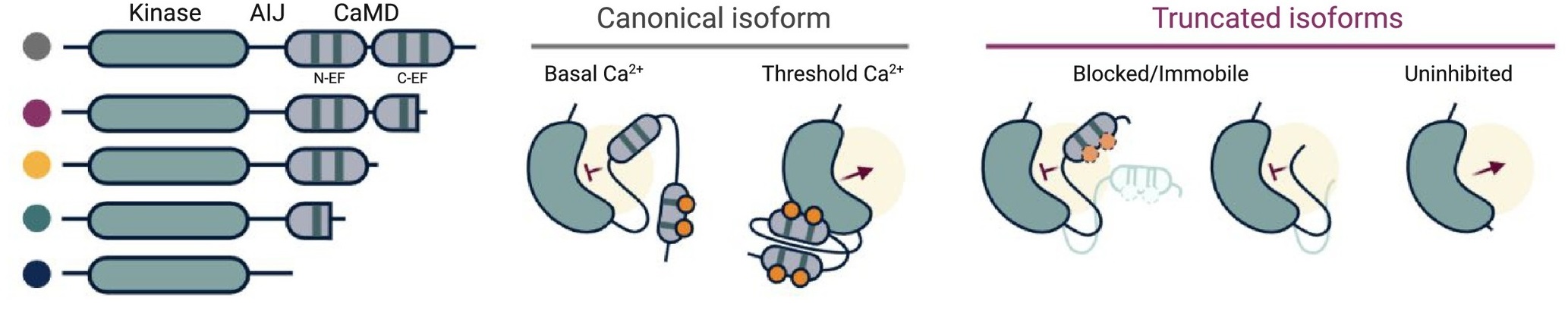 Calcium-dependent protein kinases (CDPKs/CPKs) are an interesting class of proteins present in plants, algae and some protists that are thought to “sense” and “respond” to spikes in intracellular Ca2+ signaling events. While multiple mechanisms have been proposed to be involved in the regulation of CDPKs, Loranger and colleagues discuss the merits of alternative splicing (AS) as a conserved factor in regulating CDPKs. Though alternative splicing was known to increase diversity of protein isoforms, it is now established to function in specific plant stress responses. In this context, the authors focus on some recent works that show treatment of plants with defense-related peptides to result in expression of alternatively spliced forms of CPK28, a protein that works at the intersection of pathogen defense and flowering transition. Through analysis of publicly available transcriptome and protein sequence data of CDPK families across angiosperms, the authors find alternatively spliced forms of CDPKs to be prevalently expressed. Notably, the truncations are seen in the CaM-like domain (CaMD) region that binds to Ca2+ ions and is thought to serve as the “sensor” domain. The authors thus propose that alternative splicing can be a “broadly-used regulatory mechanism” in plants to express CaMD-truncated isoforms of CDPKs in response to pathogen attack. Though the functional relevance of such a regulatory mechanism is yet to be demonstrated, it would also be useful in understanding the role of Ca2+ binding in activation and functioning of CDPKs. (Summary by Pavithran Narayanan @pavi_narayanan) Trends Plant Sci. 10.1016/j.tplants.2021.07.002
Calcium-dependent protein kinases (CDPKs/CPKs) are an interesting class of proteins present in plants, algae and some protists that are thought to “sense” and “respond” to spikes in intracellular Ca2+ signaling events. While multiple mechanisms have been proposed to be involved in the regulation of CDPKs, Loranger and colleagues discuss the merits of alternative splicing (AS) as a conserved factor in regulating CDPKs. Though alternative splicing was known to increase diversity of protein isoforms, it is now established to function in specific plant stress responses. In this context, the authors focus on some recent works that show treatment of plants with defense-related peptides to result in expression of alternatively spliced forms of CPK28, a protein that works at the intersection of pathogen defense and flowering transition. Through analysis of publicly available transcriptome and protein sequence data of CDPK families across angiosperms, the authors find alternatively spliced forms of CDPKs to be prevalently expressed. Notably, the truncations are seen in the CaM-like domain (CaMD) region that binds to Ca2+ ions and is thought to serve as the “sensor” domain. The authors thus propose that alternative splicing can be a “broadly-used regulatory mechanism” in plants to express CaMD-truncated isoforms of CDPKs in response to pathogen attack. Though the functional relevance of such a regulatory mechanism is yet to be demonstrated, it would also be useful in understanding the role of Ca2+ binding in activation and functioning of CDPKs. (Summary by Pavithran Narayanan @pavi_narayanan) Trends Plant Sci. 10.1016/j.tplants.2021.07.002
Structural and biochemical analyses of concanavalin A circular permutation
 If you have heard of concanavalin A (ConA), it probably is because it is a widely-used reagent in carbohydrate science and medical research. ConA is a lectin derived from jackbean (Canavalia ensiformis, hence the name) that binds assorted carbohydrates. It is also a fascinating protein that assembles in a unique way. Post-translationally, the pro-protein is cleaved in the middle, the front and back half sides switch and then are rejoined by the formation of a new peptide bond – this is a type of circular permutation (check out the Wikipedia page on Circular Permutation in Proteins). This sort of rearrangement usually occurs at the genetic level, or by fusion of two different proteins; ConA is unique in that it occurs post-translationally to a single polypeptide. Here, Nonis et al. expressed recombinant pro-ConA and the enzyme that cuts and pastes it, asparaginyl endopeptidase (CeAEP1), and solved crystal structures for the pro-ConA, ConA, and CeAEP1. Their analysis shows that the rearrangement leads to greater protein stability. This work is more than an investigation into an evolutionary curiosity, it also provides insights that can be applied to protein engineering. (Summary by Mary Williams @PlantTeaching) Plant Cell 10.1093/plcell/koab130
If you have heard of concanavalin A (ConA), it probably is because it is a widely-used reagent in carbohydrate science and medical research. ConA is a lectin derived from jackbean (Canavalia ensiformis, hence the name) that binds assorted carbohydrates. It is also a fascinating protein that assembles in a unique way. Post-translationally, the pro-protein is cleaved in the middle, the front and back half sides switch and then are rejoined by the formation of a new peptide bond – this is a type of circular permutation (check out the Wikipedia page on Circular Permutation in Proteins). This sort of rearrangement usually occurs at the genetic level, or by fusion of two different proteins; ConA is unique in that it occurs post-translationally to a single polypeptide. Here, Nonis et al. expressed recombinant pro-ConA and the enzyme that cuts and pastes it, asparaginyl endopeptidase (CeAEP1), and solved crystal structures for the pro-ConA, ConA, and CeAEP1. Their analysis shows that the rearrangement leads to greater protein stability. This work is more than an investigation into an evolutionary curiosity, it also provides insights that can be applied to protein engineering. (Summary by Mary Williams @PlantTeaching) Plant Cell 10.1093/plcell/koab130
Plant PIEZO homologs modulate vacuole morphology during tip growth
 The ability to perceive and respond to mechanical stimuli and forces, such as gravity or touch, is an extremely well conserved property that is key for proper cellular function. This ability is fueled by mechanosensitive ion channels activated by mechanical disturbances in the membrane, which leads to flow of ions down their electrochemical gradients. In animals, the PIEZO family of mechanosensitive channels located in the plasma membrane conduct calcium and are necessary for light touch perception, compressive force, proprioception, nociception, among other processes. Interestingly, although PIEZO homologs can be found in genomes across the eukaryotic tree of life, the biomechanics of the plant cell imposed by its cell wall pose an interesting question: how might PIEZO channels function in plants? Seeking to answer this question, Radin and colleagues recently investigated the PIEZO homologs of the moss Physcomitrium patens (PpPIEZOs) and Arabidopsis thaliana (AtPIEZO) using different types of mutants (loss-of-function [LOF], gain-of-function, overexpression), reporter lines and vacuolar structure-specific dyes. The authors discovered that PpPIEZO proteins localize to the vacuole membrane (tonoplast) in subapical and apical caulonemal cells and are required for proper growth and cytoplasmic Ca2+ oscillation signatures. In the wild-type, caulonemal cells contain tubulated and fragmented vacuoles, in contrast, the vacuoles in the LOF mutant background are fused and expanded, therefore suggesting that PpPIEZOs are necessary for normal tubulated vacuoles. The authors were able to recapitulate the LOF phenotype using a gain-of-function mutant in which a key amino acid residue was deleted, thus further supporting their conclusions: PpPIEZOs influence vacuolar fission/morphology. Finally, the authors assessed the conservation of the mechanism of PIEZO proteins mechanism by AtPIEZO in moss: indeed, AtPIEZO localizes to the tonoplast and suppresses the vacuolar defects observed in the PpPIEZO LOF background. Moreover, AtPIEZO LOF mutants show similar vacuolar defects as the ones observed in moss, therefore suggesting that the role of PIEZO proteins in promoting vacuolar fission and invagination is conserved between moss and Arabidopsis. This interesting work spotlights how PIEZO plant homologs may have been co-opted to sense the mechanical of the tonoplast, rather than of the plasma membrane like in animals. (Summary by Jesus Leon @jesussaur) Science 10.1126/science.abe6310
The ability to perceive and respond to mechanical stimuli and forces, such as gravity or touch, is an extremely well conserved property that is key for proper cellular function. This ability is fueled by mechanosensitive ion channels activated by mechanical disturbances in the membrane, which leads to flow of ions down their electrochemical gradients. In animals, the PIEZO family of mechanosensitive channels located in the plasma membrane conduct calcium and are necessary for light touch perception, compressive force, proprioception, nociception, among other processes. Interestingly, although PIEZO homologs can be found in genomes across the eukaryotic tree of life, the biomechanics of the plant cell imposed by its cell wall pose an interesting question: how might PIEZO channels function in plants? Seeking to answer this question, Radin and colleagues recently investigated the PIEZO homologs of the moss Physcomitrium patens (PpPIEZOs) and Arabidopsis thaliana (AtPIEZO) using different types of mutants (loss-of-function [LOF], gain-of-function, overexpression), reporter lines and vacuolar structure-specific dyes. The authors discovered that PpPIEZO proteins localize to the vacuole membrane (tonoplast) in subapical and apical caulonemal cells and are required for proper growth and cytoplasmic Ca2+ oscillation signatures. In the wild-type, caulonemal cells contain tubulated and fragmented vacuoles, in contrast, the vacuoles in the LOF mutant background are fused and expanded, therefore suggesting that PpPIEZOs are necessary for normal tubulated vacuoles. The authors were able to recapitulate the LOF phenotype using a gain-of-function mutant in which a key amino acid residue was deleted, thus further supporting their conclusions: PpPIEZOs influence vacuolar fission/morphology. Finally, the authors assessed the conservation of the mechanism of PIEZO proteins mechanism by AtPIEZO in moss: indeed, AtPIEZO localizes to the tonoplast and suppresses the vacuolar defects observed in the PpPIEZO LOF background. Moreover, AtPIEZO LOF mutants show similar vacuolar defects as the ones observed in moss, therefore suggesting that the role of PIEZO proteins in promoting vacuolar fission and invagination is conserved between moss and Arabidopsis. This interesting work spotlights how PIEZO plant homologs may have been co-opted to sense the mechanical of the tonoplast, rather than of the plasma membrane like in animals. (Summary by Jesus Leon @jesussaur) Science 10.1126/science.abe6310
The secret to leaf forever: Mechanisms controlling simple leaf development
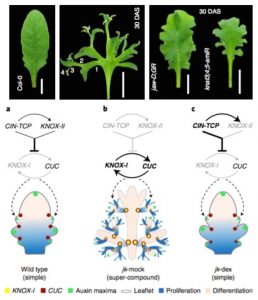 The final shape of a leaf is a consequence of differential growth at its margins. Simple (or lightly serrated) leaves result from limited growth in the margins, while compound leaves result from a constant initiation of leaflets. A recent work by Challa, Rath, and colleagues dissected the regulatory network controlling leaf shape. Two modules control the growth at the leaf margin, one involving CINCINNATA-like TCP (CIN-TCP) and the other that includes some class-II KNOTTED1-LIKE (KNOX-II) transcription factors. Mutants affected in any of these modules display deep serrations in the leaves. The combined suppression of the CIN-TPC and KNOX-II modules causes the continued emergence of leaflets, resembling a compound leaf. In this background, the inactivation of both CIN-TCP and the KNOX-II modules allows the enhanced activity of KNOX-I genes (KNAT2, KNAT6) and CUC2. This circuit restores meristematic features in the leaf margins and causes the emergence of leaflets. The absence of the suppression modules confers indeterminate growth features to the leaves. (Summary by Humberto Herrera-Ubaldo @herrera_h) Nature Plants 10.1038/s41477-021-00965-3
The final shape of a leaf is a consequence of differential growth at its margins. Simple (or lightly serrated) leaves result from limited growth in the margins, while compound leaves result from a constant initiation of leaflets. A recent work by Challa, Rath, and colleagues dissected the regulatory network controlling leaf shape. Two modules control the growth at the leaf margin, one involving CINCINNATA-like TCP (CIN-TCP) and the other that includes some class-II KNOTTED1-LIKE (KNOX-II) transcription factors. Mutants affected in any of these modules display deep serrations in the leaves. The combined suppression of the CIN-TPC and KNOX-II modules causes the continued emergence of leaflets, resembling a compound leaf. In this background, the inactivation of both CIN-TCP and the KNOX-II modules allows the enhanced activity of KNOX-I genes (KNAT2, KNAT6) and CUC2. This circuit restores meristematic features in the leaf margins and causes the emergence of leaflets. The absence of the suppression modules confers indeterminate growth features to the leaves. (Summary by Humberto Herrera-Ubaldo @herrera_h) Nature Plants 10.1038/s41477-021-00965-3
A slicing mechanism facilitates host entry by plant-pathogenic Phytophthora
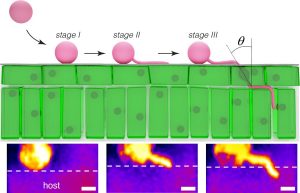 A plant has many layers of defense against a pathogen. One of the first challenges a pathogen faces is how to get inside the plant. Some bacteria sneak in through open stomatal pores, and some fungi form high-pressure appressoria that burst through walls. Here, Bronkhorst et al. investigated how the Phytophthora oomycete pathogens get into a plant. The authors noted that, unlike a fungal appressorium that penetrates at a 90° angle into the tissue, the oomycyte hypha enters on a diagonal. This type of oblique indentation is known in mechanics studies as slicing. Using an artificial, cuticle-like substrate and a fluorogenic mechanosensor, the authors investigated this entry mechanism, which they refer to as “naifu” (Japanese for knife). They found that it requires cell polarization involving the actin cytoskeleton, substrate adherence through protein secretions, and turgor generation. This new understanding of how Phytophthora first breach plant defenses suggests targets for control of these destructive plant killers. (Summary by Mary Williams @PlantTeaching) Nature Microbiol. 10.1038/s41564-021-00919-7
A plant has many layers of defense against a pathogen. One of the first challenges a pathogen faces is how to get inside the plant. Some bacteria sneak in through open stomatal pores, and some fungi form high-pressure appressoria that burst through walls. Here, Bronkhorst et al. investigated how the Phytophthora oomycete pathogens get into a plant. The authors noted that, unlike a fungal appressorium that penetrates at a 90° angle into the tissue, the oomycyte hypha enters on a diagonal. This type of oblique indentation is known in mechanics studies as slicing. Using an artificial, cuticle-like substrate and a fluorogenic mechanosensor, the authors investigated this entry mechanism, which they refer to as “naifu” (Japanese for knife). They found that it requires cell polarization involving the actin cytoskeleton, substrate adherence through protein secretions, and turgor generation. This new understanding of how Phytophthora first breach plant defenses suggests targets for control of these destructive plant killers. (Summary by Mary Williams @PlantTeaching) Nature Microbiol. 10.1038/s41564-021-00919-7
Relevance of hop terroir for beer flavor
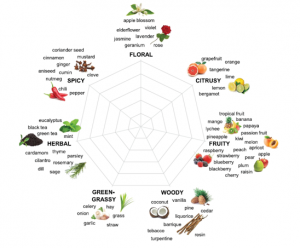 Beer is a simple beverage, basically fermented barley sugars. But recently, it’s been all about the hops. Hops are buds of Humulus lupulus, a member of the Cannabaceae family, and from humble origins they have been carefully bred and selected, resulting in many varieties with different aroma and flavor characteristics. “Terroir” is a term usually associated with wines, and refers to how the environment in which grapes are grown, from soil types to rainfall and temperature, affects the flavor of the wine. Here, Van Holle et al. explore the relevance of terroir for beer flavor. Not unexpectedly, the authors found that growing region affects the characteristics of beer made from genetically identical hops varieties – a classic example of the gene x environment (GxE) interaction effects on hop chemistry. Given the tremendous recent growth in beer connoisseurship, I wonder if we won’t soon be seeing vintages associated with our favorite craft beers. (Summary by Mary Williams @PlantTeaching) J. Inst. Brew. 10.1002/jib.648
Beer is a simple beverage, basically fermented barley sugars. But recently, it’s been all about the hops. Hops are buds of Humulus lupulus, a member of the Cannabaceae family, and from humble origins they have been carefully bred and selected, resulting in many varieties with different aroma and flavor characteristics. “Terroir” is a term usually associated with wines, and refers to how the environment in which grapes are grown, from soil types to rainfall and temperature, affects the flavor of the wine. Here, Van Holle et al. explore the relevance of terroir for beer flavor. Not unexpectedly, the authors found that growing region affects the characteristics of beer made from genetically identical hops varieties – a classic example of the gene x environment (GxE) interaction effects on hop chemistry. Given the tremendous recent growth in beer connoisseurship, I wonder if we won’t soon be seeing vintages associated with our favorite craft beers. (Summary by Mary Williams @PlantTeaching) J. Inst. Brew. 10.1002/jib.648



