Plant Science Research Weekly: August 13, 2021
A Central Role for Genetics in Plant Biology

(Left to right) Shauna and Chris Somerville, Elliot Meyerowitz, David Meinke, Marta Crouch, and Maarten Koornneef in 1985.
Plant biology as a field of study encompasses a rich plethora of subjects, some of which themselves are their own fields. Maarten Koornneef, in this memoir-style article, describes how genetics in particular shaped his career that saw the rise of Arabidopsis as the lovable poster child of plant biology today. Starting with a background in plant breeding allowed Koornneef to provide valuable insight in the beginnings of many different projects. For instance, Koornneef’s keen eye for plant traits allowed him to identify important phenotypes from mutagenic screens, leading to the isolation of several plant hormone mutants, as well as the development of genetic maps and QTL analysis for Arabidopsis. Genetics work in future projects allowed for key developments in tomato cell culture technologies and the pioneering use of natural variation in Arabidopsis accessions to further genetic and ecological evolution studies. Alongside the many projects involved in these diverse topics, Koornneef describes the important collaborations with other geneticists, molecular biologists, computational biologists, and plant biology institutions that made plant biology and Arabidopsis research as fruitful as it is today. He concludes: “I realize that during my career I was part of one of the most exciting periods in plant science, in which plant biology became one topic instead of a group of multiple disciplines.” (Summary by Benjamin Jin) Annu. Rev. Plant Biol. 10.1146/annurev-arplant-071720-111039
A research road map for responsible use of agricultural nitrogen
 Recently, the UN Environment Programme identified excessive reactive nitrogen (N) resulting from anthropogenic activity as one of the five emerging threats facing our planet. Much of this derives from agricultural practices in which N fertilizers are inefficiently applied to crops, a significant proportion of N being lost to surrounding environments. Udvardi et al. outline a roadmap for future years in how we can optimize agricultural N use efficiency (NUE) over multiple N-related practices. This roadmap can be summarized temporally. In the short-term, focusing on improving agricultural practices, such as better synchronizing N deployment with a crop’s actual N-usage, would help decrease N-pollution and inform future practices. Over time, these practices can be optimized with multiple N-related R&D discoveries, including plant breeding for more N-efficient cultivars, or improved utilization of N-fixers in legumes or potentially even non-legumes. Alongside a plethora of reviewed methodologies for improving NUE, the authors note that in addition to scientific solutions, there is a societal role necessary for solving our global issue of N-pollution, including “…expanding the farming objectives from the primary focus on production-for-profit… to include stewardship-of-the-land and surrounding environment, which is crucial for sustainability of agriculture and society.” (Summary by Benjamin Jin) Front. Sustain. Food Syst. 10.3389/fsufs.2021.660155
Recently, the UN Environment Programme identified excessive reactive nitrogen (N) resulting from anthropogenic activity as one of the five emerging threats facing our planet. Much of this derives from agricultural practices in which N fertilizers are inefficiently applied to crops, a significant proportion of N being lost to surrounding environments. Udvardi et al. outline a roadmap for future years in how we can optimize agricultural N use efficiency (NUE) over multiple N-related practices. This roadmap can be summarized temporally. In the short-term, focusing on improving agricultural practices, such as better synchronizing N deployment with a crop’s actual N-usage, would help decrease N-pollution and inform future practices. Over time, these practices can be optimized with multiple N-related R&D discoveries, including plant breeding for more N-efficient cultivars, or improved utilization of N-fixers in legumes or potentially even non-legumes. Alongside a plethora of reviewed methodologies for improving NUE, the authors note that in addition to scientific solutions, there is a societal role necessary for solving our global issue of N-pollution, including “…expanding the farming objectives from the primary focus on production-for-profit… to include stewardship-of-the-land and surrounding environment, which is crucial for sustainability of agriculture and society.” (Summary by Benjamin Jin) Front. Sustain. Food Syst. 10.3389/fsufs.2021.660155
Large-scale whole-genome resequencing unravels the domestication history of Cannabis sativa
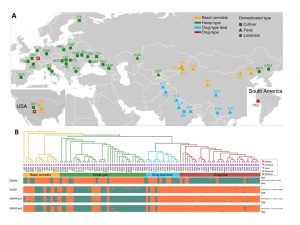 Throughout history, many cultures have exploited the properties of Cannabis sativa, either as a source of fiber extracts (hemp) or as a medicinal and recreational drug (drug type). It is thought that cannabis was originally cultivated in Asia from where it spread to other regions around the globe, and it was during this process that artificial selection and outcrossing with other cultivars or landraces occurred. Cannabis produce two main types of secondary metabolites: cannabidiolic acid (CBDA) and Δ9-tetrahydrocannabinolic acid (THCA). These compounds are of medical importance because they bind to endocannabinoid receptors in an animal’s nervous system and could treat some neurological disorders. Cannabis hemp, drug types, and hybrid cultivars differ in their relative yield of these compounds and have been classified according to their chemotype. Despite the long history of cannabis use and the need to produce varieties with specific cannabinoid profiles, the genomic history of domestication and the mechanisms mediating the evolution of cannabinoid synthase genes has been understudied. Here, the authors investigated C. sativa domestication history by analyzing whole-genome profiles of 110 accessions distributed around the world. They showed that C. sativa was first domesticated in early Neolithic times in East Asia. Phylogenetic analysis showed a strong clustering of Cannabis accessions into four well-separated genetic groups [A) Basal, B) Hemp-type, C) Drug-type feral and D) Drug-type] and suggest that all current hemp and drug cultivars diverged from an ancestral gene pool represented by feral plants and landraces in China. Additionally, the authors identified a set of specific candidate genes that appeared to have been under selection differentiating hemp and drug types. Such genes affected traits that where expected to have been under selection, such as branching pattern and cellulose/lignin biosynthesis, leading to unbranched, cellulose-rich/lignin-poor bast fibers in hemp types versus well-branched with lignin-rich woody cores, maximizing flower and resin production in drug types. Interestingly, the authors also found that genes involved in the synthesis of the two major biochemically competing cannabinoids THCAS or CBDAS were lost during domestication for increased fiber production or psychoactive properties respectively. In conclusion, this study provides insights into the domestication history of C. sativa and identifies putative genes that have been under selection, supporting an evolutionary scenario that accounts for the variability in cannabinoid composition. Furthermore, the study offers an unprecedented base of genomic resources for breeders for both medical and agricultural applications. (Summary by Daniela Ramos @DanielaR_cruz) Sci. Adv. 10.1126/sciadv.abg2286
Throughout history, many cultures have exploited the properties of Cannabis sativa, either as a source of fiber extracts (hemp) or as a medicinal and recreational drug (drug type). It is thought that cannabis was originally cultivated in Asia from where it spread to other regions around the globe, and it was during this process that artificial selection and outcrossing with other cultivars or landraces occurred. Cannabis produce two main types of secondary metabolites: cannabidiolic acid (CBDA) and Δ9-tetrahydrocannabinolic acid (THCA). These compounds are of medical importance because they bind to endocannabinoid receptors in an animal’s nervous system and could treat some neurological disorders. Cannabis hemp, drug types, and hybrid cultivars differ in their relative yield of these compounds and have been classified according to their chemotype. Despite the long history of cannabis use and the need to produce varieties with specific cannabinoid profiles, the genomic history of domestication and the mechanisms mediating the evolution of cannabinoid synthase genes has been understudied. Here, the authors investigated C. sativa domestication history by analyzing whole-genome profiles of 110 accessions distributed around the world. They showed that C. sativa was first domesticated in early Neolithic times in East Asia. Phylogenetic analysis showed a strong clustering of Cannabis accessions into four well-separated genetic groups [A) Basal, B) Hemp-type, C) Drug-type feral and D) Drug-type] and suggest that all current hemp and drug cultivars diverged from an ancestral gene pool represented by feral plants and landraces in China. Additionally, the authors identified a set of specific candidate genes that appeared to have been under selection differentiating hemp and drug types. Such genes affected traits that where expected to have been under selection, such as branching pattern and cellulose/lignin biosynthesis, leading to unbranched, cellulose-rich/lignin-poor bast fibers in hemp types versus well-branched with lignin-rich woody cores, maximizing flower and resin production in drug types. Interestingly, the authors also found that genes involved in the synthesis of the two major biochemically competing cannabinoids THCAS or CBDAS were lost during domestication for increased fiber production or psychoactive properties respectively. In conclusion, this study provides insights into the domestication history of C. sativa and identifies putative genes that have been under selection, supporting an evolutionary scenario that accounts for the variability in cannabinoid composition. Furthermore, the study offers an unprecedented base of genomic resources for breeders for both medical and agricultural applications. (Summary by Daniela Ramos @DanielaR_cruz) Sci. Adv. 10.1126/sciadv.abg2286
Plant scientists’ research attention is skewed towards colourful, conspicuous and broadly distributed flowers
 Chicana feminist writer Cherríe Moraga wrote in her anthology This Bridge Called My Back, with regards to the necessary inclusion of women of color in contemporary progressive politics: “It is not always a matter of the actual bodies in the room, but of a life dedicated to a growing awareness of who and what is missing in that room” — this sentiment, describing an awareness of what (or who) is ignored by our implicit biases, is vital in all fields of research/activism. This includes plant biology and conservation. Concepts such as ‘plant awareness disparity’ highlight a relative lack of awareness for plants in biological research, but even within plant research there may be unidentified biases in how certain plant species are selected for study. To determine if biases exist, Adamo et al. focused upon a model system of 113 plant species studied in the Southwestern Alps and performed bibliometrics/trait-based approaches on publications relating to these species. Initial research suggested that morphology and colour were more pertinent traits towards research attention than ecology and rarity. Further research revealed significant relationships between a species’ selection for research and flower colour (particularly striking colours that contrast a brown/green environmental background); plant stem height (a taller plant implies greater conspicuousness); and range size (plants with a broader distribution are perhaps more accessible, thus receiving greater attention). The authors conclude that this data suggests an aesthetic bias in plant research that needs scrutiny for better research practices. (Summary by Benjamin Jin) Nature Plants 10.1038/s41477-021-00912-2
Chicana feminist writer Cherríe Moraga wrote in her anthology This Bridge Called My Back, with regards to the necessary inclusion of women of color in contemporary progressive politics: “It is not always a matter of the actual bodies in the room, but of a life dedicated to a growing awareness of who and what is missing in that room” — this sentiment, describing an awareness of what (or who) is ignored by our implicit biases, is vital in all fields of research/activism. This includes plant biology and conservation. Concepts such as ‘plant awareness disparity’ highlight a relative lack of awareness for plants in biological research, but even within plant research there may be unidentified biases in how certain plant species are selected for study. To determine if biases exist, Adamo et al. focused upon a model system of 113 plant species studied in the Southwestern Alps and performed bibliometrics/trait-based approaches on publications relating to these species. Initial research suggested that morphology and colour were more pertinent traits towards research attention than ecology and rarity. Further research revealed significant relationships between a species’ selection for research and flower colour (particularly striking colours that contrast a brown/green environmental background); plant stem height (a taller plant implies greater conspicuousness); and range size (plants with a broader distribution are perhaps more accessible, thus receiving greater attention). The authors conclude that this data suggests an aesthetic bias in plant research that needs scrutiny for better research practices. (Summary by Benjamin Jin) Nature Plants 10.1038/s41477-021-00912-2
Guard cell endomembrane Ca2+-ATPases underpin a ‘carbon memory’ of photosynthetic assimilation that impacts on water-use efficiency
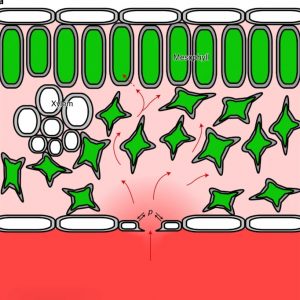 Stomatal guard cells control both carbon dioxide uptake and transpirational water loss. Guard cell control over the stomatal pore aperture is sensitive to water status (through ABA) as well as the amount of CO2 available within the leaf air space. Several studies have indicated that stomatal aperture can be improved and made more responsive for optimal water-use efficiency. Here, Jezek, Silva-Alvim, Hills, et al. combined modeling studies (using the OnGuard system) and experimental approaches to further identify guard cell control systems. They found key roles for endomembrane-localized ATP-driven calcium pumps (Ca2+-ATPases) and calcium channels, which collectively affect the concentration of cytosolic calcium ions ([Ca2+]i), which affect the other membrane transporters involved in stomatal responses. Specifically, they found that as CO2 levels fluctuate rapidly, stomatal closure responses are slowed, and this is exacerbated in mutants with deceased levels of Ca2+-ATPases. Thus, the Ca2+ stores determine a latency or ‘carbon memory’ that affects stomatal kinetics. These findings demonstrate the utility of models for identifying targets for future optimization. (Summary by Mary Williams @PlantTeaching) Nature Plants 10.1038/s41477-021-00966-2
Stomatal guard cells control both carbon dioxide uptake and transpirational water loss. Guard cell control over the stomatal pore aperture is sensitive to water status (through ABA) as well as the amount of CO2 available within the leaf air space. Several studies have indicated that stomatal aperture can be improved and made more responsive for optimal water-use efficiency. Here, Jezek, Silva-Alvim, Hills, et al. combined modeling studies (using the OnGuard system) and experimental approaches to further identify guard cell control systems. They found key roles for endomembrane-localized ATP-driven calcium pumps (Ca2+-ATPases) and calcium channels, which collectively affect the concentration of cytosolic calcium ions ([Ca2+]i), which affect the other membrane transporters involved in stomatal responses. Specifically, they found that as CO2 levels fluctuate rapidly, stomatal closure responses are slowed, and this is exacerbated in mutants with deceased levels of Ca2+-ATPases. Thus, the Ca2+ stores determine a latency or ‘carbon memory’ that affects stomatal kinetics. These findings demonstrate the utility of models for identifying targets for future optimization. (Summary by Mary Williams @PlantTeaching) Nature Plants 10.1038/s41477-021-00966-2
A prion-like protein regulator of seed germination undergoes hydration-dependent phase separation ($)
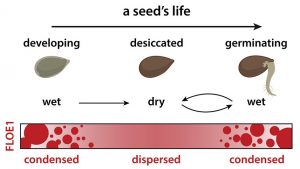 Seed imbibition (i.e., water uptake) marks the beginning of germination, an irreversible process that might lead plants to death if it occurs under unsuitable conditions for seedling development. Therefore, it is expected that seeds only germinate when water is readably available. However, the molecular mechanism behind this control remains to be described. In this fascinating research, Dorone and colleagues provide a thorough description of FLOE1, a prion-like protein that allows Arabidopsis thaliana seeds to sense water availability. FLOE1 is accumulated during embryo development and can form cytoplasmatic condensates in response to hydration. floe1 mutants had higher germination under water deficit conditions, suggesting this protein prevents germination under stressful situations. However, the loss of the function of this protein did not impact abscisic acid and gibberellin balance, a renowned regulator of germination. Overall, these results imply that FLOE1 controls germination by assessing water availability in the environment. Interestingly, the analysis of different A. thaliana ecotypes showed that FLOE1 expression was correlated with climate, indicating the importance of this protein for local adaptation. While future studies must assess the precise molecular function of FLOE1, this research provides an exciting starting point to unravel the mechanism that allows seeds to sense water. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Cell 10.1016/j.cell.2021.06.009
Seed imbibition (i.e., water uptake) marks the beginning of germination, an irreversible process that might lead plants to death if it occurs under unsuitable conditions for seedling development. Therefore, it is expected that seeds only germinate when water is readably available. However, the molecular mechanism behind this control remains to be described. In this fascinating research, Dorone and colleagues provide a thorough description of FLOE1, a prion-like protein that allows Arabidopsis thaliana seeds to sense water availability. FLOE1 is accumulated during embryo development and can form cytoplasmatic condensates in response to hydration. floe1 mutants had higher germination under water deficit conditions, suggesting this protein prevents germination under stressful situations. However, the loss of the function of this protein did not impact abscisic acid and gibberellin balance, a renowned regulator of germination. Overall, these results imply that FLOE1 controls germination by assessing water availability in the environment. Interestingly, the analysis of different A. thaliana ecotypes showed that FLOE1 expression was correlated with climate, indicating the importance of this protein for local adaptation. While future studies must assess the precise molecular function of FLOE1, this research provides an exciting starting point to unravel the mechanism that allows seeds to sense water. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Cell 10.1016/j.cell.2021.06.009
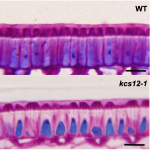
A seed coat-specific β-ketoacyl-CoA synthase, KCS12, is critical for preserving seed physical dormancy ($)
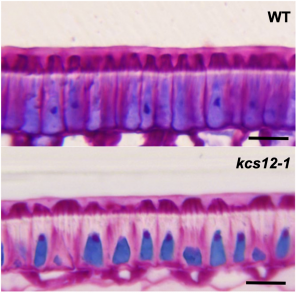 Physical dormancy is a dormancy class caused by a water-impermeable cell layer that prevents seed water uptake. Despite being present in several families, its molecular basis has seldom been addressed. Here, Chai and colleagues identify a β-ketoacyl-CoA synthase (KCS) –one of the enzymes that participate in very long chain fatty acid (VLCFA) biosynthesis– involved in the acquisition of physical dormancy in Medicago truncatula. Previous studies reported that M. truncatula knox4 mutants lack physical dormancy and presented less content of VLCFAs. Moreover, these mutants also exhibited a downregulation in KCS12, a KCS whose expression was specific to the seed coat. Further mutant analysis showed that kcs12 seeds were not physically dormant and could absorb water just like scarified wild-type seeds. In line with these, kcs12 seeds had a thinner cuticle with altered VLCFAs composition. Notably, these seeds showed a reduction in the content of 24C VLCFAs, suggesting that KCS12 is responsible for their synthesis and that they are required to achieve physical dormancy. Therefore, this research provides exciting insights into the molecular mechanisms underpinning physical dormancy. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Plant Physiol. 10.1093/plphys/kiab152
Physical dormancy is a dormancy class caused by a water-impermeable cell layer that prevents seed water uptake. Despite being present in several families, its molecular basis has seldom been addressed. Here, Chai and colleagues identify a β-ketoacyl-CoA synthase (KCS) –one of the enzymes that participate in very long chain fatty acid (VLCFA) biosynthesis– involved in the acquisition of physical dormancy in Medicago truncatula. Previous studies reported that M. truncatula knox4 mutants lack physical dormancy and presented less content of VLCFAs. Moreover, these mutants also exhibited a downregulation in KCS12, a KCS whose expression was specific to the seed coat. Further mutant analysis showed that kcs12 seeds were not physically dormant and could absorb water just like scarified wild-type seeds. In line with these, kcs12 seeds had a thinner cuticle with altered VLCFAs composition. Notably, these seeds showed a reduction in the content of 24C VLCFAs, suggesting that KCS12 is responsible for their synthesis and that they are required to achieve physical dormancy. Therefore, this research provides exciting insights into the molecular mechanisms underpinning physical dormancy. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Plant Physiol. 10.1093/plphys/kiab152