Plant Science Research Weekly: April 29, 2022
Review: Fire-released seed dormancy – a global synthesis
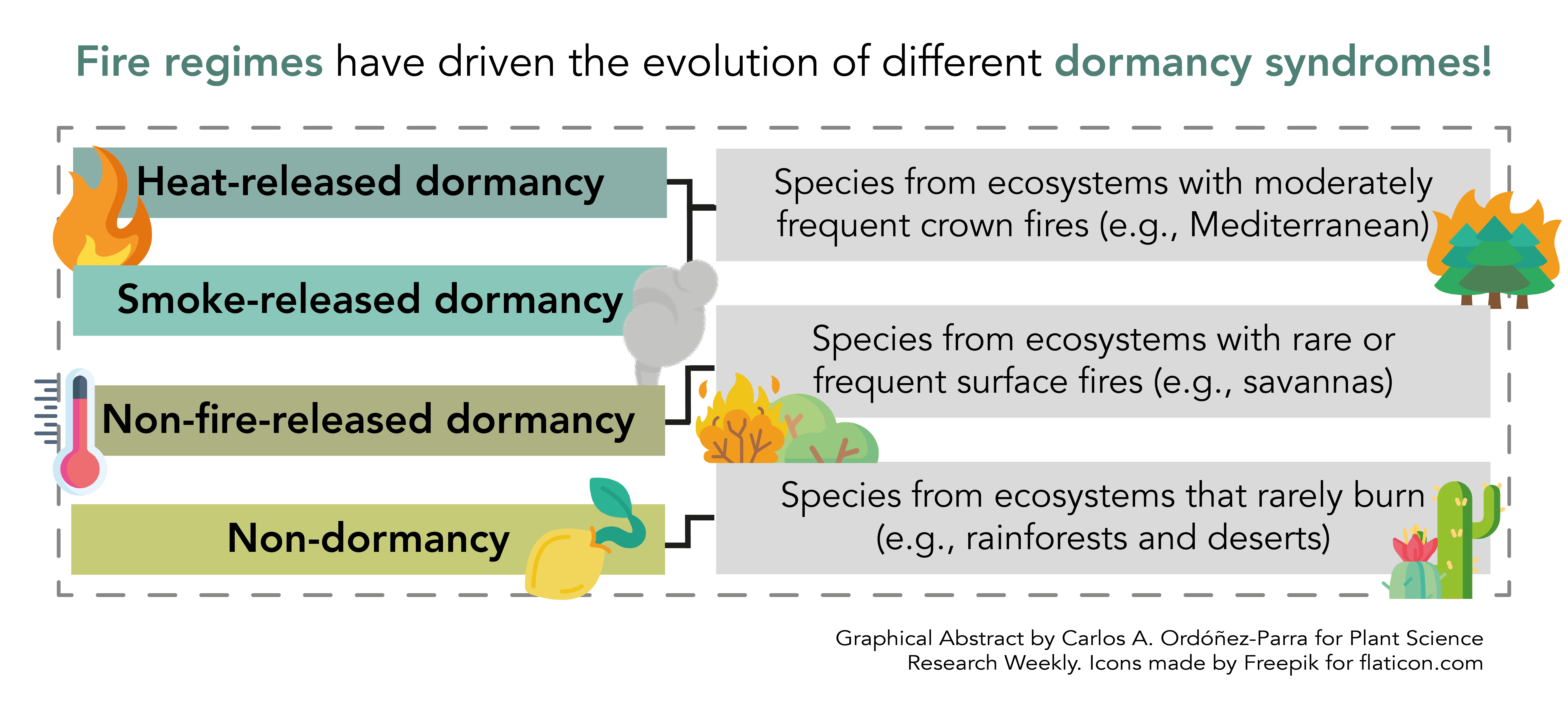 In 1991, Jon E. Keely wrote the first review about the role of fire as a dormancy-breaking cue in California’s Mediterranean ecosystems. In this review, almost 30 years after Keely’s work, Pausas and Lamont provide us with an updated and worldwide vision of the subject. Four dormancy syndromes and their potential evolutionary drivers are presented. Heat- and smoked-released dormancy are typical in ecosystems subject to frequent crown fires like Mediterranean ones. These dormancy syndromes have evolved several independent times during plant evolution, potentially during the Cretaceous, where fires were fairly common, and due to species migration from non-fireprone ecosystems to fireprone ones. In contrast, species from ecosystems where fires are rarer and superficial have developed dormancy systems that are not broken by fire but by other environmental factors, such as moisture and warmth. Finally, species from ecosystems where fires are extremely rare –such as rainforests– have many species with non-dormant seeds. While it highlights other environmental factors besides fire that might have driven the evolution of dormancy in fireprone ecosystems, this review highlights how fire and its regimes set the scene for it. Would you like to know more about this fiery topic? This detailed and compelling reading is an excellent place to start. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Biol. Rev. 10.1111/brv.12855
In 1991, Jon E. Keely wrote the first review about the role of fire as a dormancy-breaking cue in California’s Mediterranean ecosystems. In this review, almost 30 years after Keely’s work, Pausas and Lamont provide us with an updated and worldwide vision of the subject. Four dormancy syndromes and their potential evolutionary drivers are presented. Heat- and smoked-released dormancy are typical in ecosystems subject to frequent crown fires like Mediterranean ones. These dormancy syndromes have evolved several independent times during plant evolution, potentially during the Cretaceous, where fires were fairly common, and due to species migration from non-fireprone ecosystems to fireprone ones. In contrast, species from ecosystems where fires are rarer and superficial have developed dormancy systems that are not broken by fire but by other environmental factors, such as moisture and warmth. Finally, species from ecosystems where fires are extremely rare –such as rainforests– have many species with non-dormant seeds. While it highlights other environmental factors besides fire that might have driven the evolution of dormancy in fireprone ecosystems, this review highlights how fire and its regimes set the scene for it. Would you like to know more about this fiery topic? This detailed and compelling reading is an excellent place to start. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Biol. Rev. 10.1111/brv.12855
Opinion. Seed traits and phylogenomics: prospects for the 21st century ($)
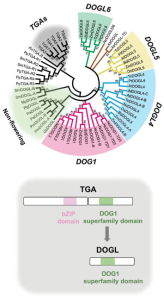 In August 2021, The International Seed Science Society hosted its 13th International Conference. The meeting had many exciting presentations, including the keynote lecture given by Dr. Hiroyuki Nonogaki, entitled “Seed traits and pylogenomics: perspectives for the 21st Century”. This paper synthesizes the presentation’s main points, providing a solid case for the use of phylogenomics –the intersection between phylogenetic methods and genomics– in the reconstruction of species’ evolutionary history. To illustrate their case, the authors present phylogenomic evidence about the evolution of the DELAY OF GERMINATION1 (DOG1) gene family, widely known for controlling seed dormancy, reserve accumulation, and desiccation tolerance. Members of this gene family can be traced back to the Zygnematophyceae, the sister algal lineage of land plants. Species from this group also have ancestral forms of genes that participate in seed dormancy and interact with DOG1, such as PYR1-LIKE. Therefore, phylogenomics promise to make use of this exciting scenario and provide fascinating insights into the evolution of the seed program. Still, the applications of phylogenomics go beyond the description of gene evolution, as it can guide future genetic studies and the development of crop technologies. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Seed Sci. Res. 10.1017/S0960258522000046
In August 2021, The International Seed Science Society hosted its 13th International Conference. The meeting had many exciting presentations, including the keynote lecture given by Dr. Hiroyuki Nonogaki, entitled “Seed traits and pylogenomics: perspectives for the 21st Century”. This paper synthesizes the presentation’s main points, providing a solid case for the use of phylogenomics –the intersection between phylogenetic methods and genomics– in the reconstruction of species’ evolutionary history. To illustrate their case, the authors present phylogenomic evidence about the evolution of the DELAY OF GERMINATION1 (DOG1) gene family, widely known for controlling seed dormancy, reserve accumulation, and desiccation tolerance. Members of this gene family can be traced back to the Zygnematophyceae, the sister algal lineage of land plants. Species from this group also have ancestral forms of genes that participate in seed dormancy and interact with DOG1, such as PYR1-LIKE. Therefore, phylogenomics promise to make use of this exciting scenario and provide fascinating insights into the evolution of the seed program. Still, the applications of phylogenomics go beyond the description of gene evolution, as it can guide future genetic studies and the development of crop technologies. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Seed Sci. Res. 10.1017/S0960258522000046
PDX1.1-dependent vitamin B6 synthesis alleviates ammonium toxicity-associated ROS production
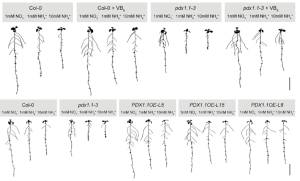 Ammonium is a nitrogen form preferred by many plant species. However, high ammonium concentrations lead to decreased primary root elongation due to membrane depolarization and lower apoplastic pH. Liu et al. report that ammonium toxicity results in iron-dependent reactive oxygen species (ROS) production and thereby decreases primary root elongation. Based on ammonium responsive transcriptome data, T-DNA mutants of 29 ammonium responsive genes were evaluated, and the T-DNA mutant of PDX1.1, a gene associated with biosynthesis of vitamin B6, was found to be hypersensitive to ammonium toxicity. Expression of PDX1.1 was upregulated by hydrogen peroxide generated by high ammonium and low pH conditions. Like other vitamins, vitamin B6 is a collection of related structures called vitamers. PDX1.1 overexpressing lines showed higher accumulation of non-phosphorylated B6 vitamers and improved the tolerance to ammonium toxicity, as well as tolerance to phosphorus deficiency and nickel toxicity. PDX1.1 is a good candidate gene for improving plant’s tolerance to growth inhibition associated with crop cultivation in ammonium rich agriculture lands. (Summary by Lekshmy Sathee @lekshmysnair) Mol. Plant 10.1016/j.molp.2022.01.012
Ammonium is a nitrogen form preferred by many plant species. However, high ammonium concentrations lead to decreased primary root elongation due to membrane depolarization and lower apoplastic pH. Liu et al. report that ammonium toxicity results in iron-dependent reactive oxygen species (ROS) production and thereby decreases primary root elongation. Based on ammonium responsive transcriptome data, T-DNA mutants of 29 ammonium responsive genes were evaluated, and the T-DNA mutant of PDX1.1, a gene associated with biosynthesis of vitamin B6, was found to be hypersensitive to ammonium toxicity. Expression of PDX1.1 was upregulated by hydrogen peroxide generated by high ammonium and low pH conditions. Like other vitamins, vitamin B6 is a collection of related structures called vitamers. PDX1.1 overexpressing lines showed higher accumulation of non-phosphorylated B6 vitamers and improved the tolerance to ammonium toxicity, as well as tolerance to phosphorus deficiency and nickel toxicity. PDX1.1 is a good candidate gene for improving plant’s tolerance to growth inhibition associated with crop cultivation in ammonium rich agriculture lands. (Summary by Lekshmy Sathee @lekshmysnair) Mol. Plant 10.1016/j.molp.2022.01.012
Nitrogen form dependent root foraging is regulated by auxin and brassinosteroid signaling
 Root growth is highly sensitive to nitrogen (N), and affected by both the form and distribution of N. The hormone auxin is a major player in root growth response to N heterogeneity. Devi et al. report on the intricate interplay of auxin and brassinosteroid (BR) signaling in determining the root growth response to low N, to ammonium, and to nitrate. Analysis of auxin and BR signaling mutants revealed transcriptome changes in support of the observed root phenotypes. Under low N, the augmented auxin transport and signaling facilitated by BR favors root elongation. By contrast, high ammonium suppresses BR signaling, which down regulates auxin transport and signaling leading to an arrest of root elongation. Nitrate-dependent root elongation also showed a close dependence on BRI1-mediated BR signaling. Thus, N-dependent root growth is coordinately regulated by auxin and BR signaling, which ensures optimum root foraging phenotype. (Summary by Lekshmy Sathee @lekshmysnair) Plant Physiol. 10.1093/plphys/kiac157
Root growth is highly sensitive to nitrogen (N), and affected by both the form and distribution of N. The hormone auxin is a major player in root growth response to N heterogeneity. Devi et al. report on the intricate interplay of auxin and brassinosteroid (BR) signaling in determining the root growth response to low N, to ammonium, and to nitrate. Analysis of auxin and BR signaling mutants revealed transcriptome changes in support of the observed root phenotypes. Under low N, the augmented auxin transport and signaling facilitated by BR favors root elongation. By contrast, high ammonium suppresses BR signaling, which down regulates auxin transport and signaling leading to an arrest of root elongation. Nitrate-dependent root elongation also showed a close dependence on BRI1-mediated BR signaling. Thus, N-dependent root growth is coordinately regulated by auxin and BR signaling, which ensures optimum root foraging phenotype. (Summary by Lekshmy Sathee @lekshmysnair) Plant Physiol. 10.1093/plphys/kiac157
ROS homeostasis mediated by MPK4 and SUMM2 determines synergid cell death
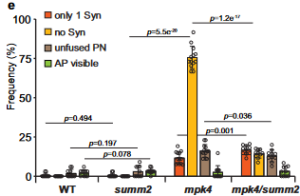 Female gametophytes (FG) of angiosperms contain eight nuclei and seven cells, distributed in a defined manner. In the micropylar end of the FG there are two synergid cells and the haploid egg, while in the central cell there are two polar nuclei. The synergid cells are responsible for attracting the pollen tube (PT). However, only one of the synergid cells will be the one that is receptive to the PT, at which point the receptive synergid instantly undergoes programmed cell death (PCD), whereas the other synergid cell persists. Mitogen-activated protein kinase (MPK) cascades are vital for plant defense; indeed, MPK4 is a negative regulator of plant immunity. The MPK4 downstream cascade is mediated by SUPPRESSOR OF mkk1 mkk2 (SUMM2) which regulates the phosphorylation of CALMODULIN-BINDING RECEPTOR LIKE KINASE 3 (CRCK3/SUMM3). In plant defense, if MPK4 is inhibited (i.e., due to a pathogenic effector), the phosphorylation of CRCK3 decreases, leading to PCD and autoimmunity. In this work, Völz et al. described the role of MPK4 in synergid preservation via the inactivation of SUMM2-mediated ROS production and subsequent cell death. The authors described that the mpk4 Arabidopsis mutant line suffers premature synergid cell death and ROS accumulation, along with an inability to reproduce. In addition, they demonstrate the localization of the MPK4 protein in FG and ovule tissue, and that by disrupting SUMM2, synergid maintenance is restored in mpk4. In conclusion, this work shows how MPK4 governs PCD of synergid cells by repressing SUMM2. (Summary by Eva Maria Gomez Alvarez, @eva_ga96). Nature Comms. 10.1038/s41467-022-29373-7
Female gametophytes (FG) of angiosperms contain eight nuclei and seven cells, distributed in a defined manner. In the micropylar end of the FG there are two synergid cells and the haploid egg, while in the central cell there are two polar nuclei. The synergid cells are responsible for attracting the pollen tube (PT). However, only one of the synergid cells will be the one that is receptive to the PT, at which point the receptive synergid instantly undergoes programmed cell death (PCD), whereas the other synergid cell persists. Mitogen-activated protein kinase (MPK) cascades are vital for plant defense; indeed, MPK4 is a negative regulator of plant immunity. The MPK4 downstream cascade is mediated by SUPPRESSOR OF mkk1 mkk2 (SUMM2) which regulates the phosphorylation of CALMODULIN-BINDING RECEPTOR LIKE KINASE 3 (CRCK3/SUMM3). In plant defense, if MPK4 is inhibited (i.e., due to a pathogenic effector), the phosphorylation of CRCK3 decreases, leading to PCD and autoimmunity. In this work, Völz et al. described the role of MPK4 in synergid preservation via the inactivation of SUMM2-mediated ROS production and subsequent cell death. The authors described that the mpk4 Arabidopsis mutant line suffers premature synergid cell death and ROS accumulation, along with an inability to reproduce. In addition, they demonstrate the localization of the MPK4 protein in FG and ovule tissue, and that by disrupting SUMM2, synergid maintenance is restored in mpk4. In conclusion, this work shows how MPK4 governs PCD of synergid cells by repressing SUMM2. (Summary by Eva Maria Gomez Alvarez, @eva_ga96). Nature Comms. 10.1038/s41467-022-29373-7
Bacterial avirulence gene encodes for a secreted protease and restricts host range
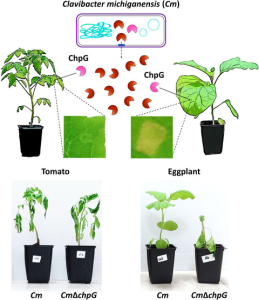 Plant pathogenic bacteria of the genus Clavibacter tend to have a narrow host range, but different species affect many important crops. Clavibacter michiganensis (Cm) causes bacterial wilt and canker in tomato, pepper and a few varieties of eggplant. There are no Cm-resistant tomato varieties but many eggplant varieties are resistant to the disease. Most Cm virulence determinants are clustered in the chp/tomA pathogenicity island (PI). Cm lacks any known translocation apparatus and therefore relies on extracellular virulence factors like hydrolases encoded in the PI. To better understand Cm host range, Verma and Teper infected tomato, pepper and eggplant with a collection of PI mutants of Cm. The authors then screened the infected plants for the hypersensitive response (HR) that is indicative of resistance. With this, the authors identified ChpG as the protein that elicits HR and is therefore recognised in many resistant varieties. ChpG had previously been reported to also trigger HR in nonhost plants. ChpG is a serine protease and its activity is required for host recognition. In fact, ChpG mutants of Cm are pathogenic on eggplant varieties that are resistant to the wild-type Cm. This suggest that this secreted protease is a determinant of host range for the bacteria. (Summary by Mariana Schuster @MariSchuster) Mol. Plant Pathol 10.1111/mpp.13215
Plant pathogenic bacteria of the genus Clavibacter tend to have a narrow host range, but different species affect many important crops. Clavibacter michiganensis (Cm) causes bacterial wilt and canker in tomato, pepper and a few varieties of eggplant. There are no Cm-resistant tomato varieties but many eggplant varieties are resistant to the disease. Most Cm virulence determinants are clustered in the chp/tomA pathogenicity island (PI). Cm lacks any known translocation apparatus and therefore relies on extracellular virulence factors like hydrolases encoded in the PI. To better understand Cm host range, Verma and Teper infected tomato, pepper and eggplant with a collection of PI mutants of Cm. The authors then screened the infected plants for the hypersensitive response (HR) that is indicative of resistance. With this, the authors identified ChpG as the protein that elicits HR and is therefore recognised in many resistant varieties. ChpG had previously been reported to also trigger HR in nonhost plants. ChpG is a serine protease and its activity is required for host recognition. In fact, ChpG mutants of Cm are pathogenic on eggplant varieties that are resistant to the wild-type Cm. This suggest that this secreted protease is a determinant of host range for the bacteria. (Summary by Mariana Schuster @MariSchuster) Mol. Plant Pathol 10.1111/mpp.13215
A plant gene that shapes its ecosystem
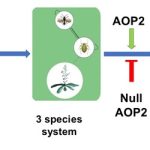
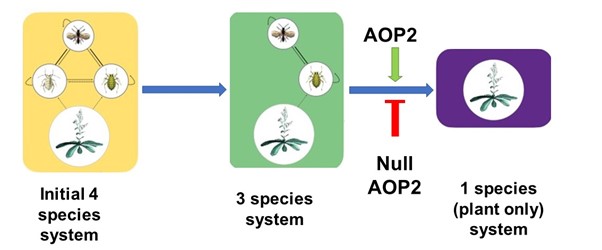 Keystone species shape the structure of an entire ecosystem, but can a gene determine structure of the whole ecosystem? A recent study indicates yes, some genes can, and such genes are called keystone genes. Barbour et al. set up a small ecosystem containing two herbivore aphids, one predator of these aphids, and Arabidopsis plants carrying mutations in any of three genes. The genes are involved in biosynthesis of glucosinolates (compounds that deter herbivory), so regulate herbivore tolerance and show natural variation in expression across various Arabidopsis genotypes. The authors observed that with time, this tiny ecosystem moved towards a three-species system with one herbivore becoming extinct. Eventually, either this three-species system remained stable, or transformed into a “plant-only” ecosystem. The authors observed one particular gene, AOP2, helps determine the final structure of this tiny ecosystem. The null allele of AOP2 helps in stabilizing the three-species food chain by increasing the plant growth rate, thus supporting growth of an aphid and its predator. Thus, Arabidopsis AOP2 acts as a keystone gene determining the structure of the ecosystem around the plant. (Summary by Kamal Kumar Malukani, @KamalMalukani) Science 10.1126/science.abf2232
Keystone species shape the structure of an entire ecosystem, but can a gene determine structure of the whole ecosystem? A recent study indicates yes, some genes can, and such genes are called keystone genes. Barbour et al. set up a small ecosystem containing two herbivore aphids, one predator of these aphids, and Arabidopsis plants carrying mutations in any of three genes. The genes are involved in biosynthesis of glucosinolates (compounds that deter herbivory), so regulate herbivore tolerance and show natural variation in expression across various Arabidopsis genotypes. The authors observed that with time, this tiny ecosystem moved towards a three-species system with one herbivore becoming extinct. Eventually, either this three-species system remained stable, or transformed into a “plant-only” ecosystem. The authors observed one particular gene, AOP2, helps determine the final structure of this tiny ecosystem. The null allele of AOP2 helps in stabilizing the three-species food chain by increasing the plant growth rate, thus supporting growth of an aphid and its predator. Thus, Arabidopsis AOP2 acts as a keystone gene determining the structure of the ecosystem around the plant. (Summary by Kamal Kumar Malukani, @KamalMalukani) Science 10.1126/science.abf2232