Plant Science Research Weekly: April 26th
Review. After the deluge: Plant revival post-flooding
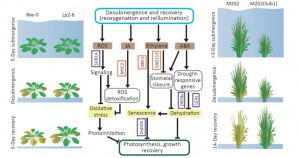 Crop losses due to flooding are occuring with increasing frequency. Whilst time submerged can be lethal, the post-submergence period is also potentially lethal. In this new review, Yeung et al. provide an overview of factors that contribute to plant survival after flooding. Being submerged leads to hypoxia (oxygen deprivation), which is then followed by re-oxygenation and exposure to light that can contribute to damaging reactive oxygen accumulation. Interestingly, damage to the root system and changes in hydraulic conductivity can lead to dessication damage after flooding. The authors explore mechanisms of flooding tolerance with a focus on rice and Arabidopsis. Tolerance draws on all aspects of plant physiology, from hormone signaling to photosynthesis and metabolism, making this an interesting and thought-provoking article. (Summary by Mary Williams) Trends Plant Sci. 10.1016/j.tplants.2019.02.007
Crop losses due to flooding are occuring with increasing frequency. Whilst time submerged can be lethal, the post-submergence period is also potentially lethal. In this new review, Yeung et al. provide an overview of factors that contribute to plant survival after flooding. Being submerged leads to hypoxia (oxygen deprivation), which is then followed by re-oxygenation and exposure to light that can contribute to damaging reactive oxygen accumulation. Interestingly, damage to the root system and changes in hydraulic conductivity can lead to dessication damage after flooding. The authors explore mechanisms of flooding tolerance with a focus on rice and Arabidopsis. Tolerance draws on all aspects of plant physiology, from hormone signaling to photosynthesis and metabolism, making this an interesting and thought-provoking article. (Summary by Mary Williams) Trends Plant Sci. 10.1016/j.tplants.2019.02.007
Review: Exchanges at the plant-oomycete interface that influence disease
 This is a great review by Judelson and Ah-Fong on the diverse signals that occur between plants and their oomycete pathogens. The authors start by describing the life- and infectivity-cycles of disease-causing oomycetes, including the familiar plant pathogens, “aggressive” Pythium and “stealthy” Phytophthora. They then cover the exchanges between host and microbe at the barrier between them. These include signals that reveal and recognize the presence of the pathogen, opportunities for pathogen entry, plant defenses and pathogen counter-defenses, nutrient exchange, and the “exit strategy” through which spores escape to start the cycle anew. The authors also expand on the formation and function of the specialized interface of the haustoria. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.18.00979
This is a great review by Judelson and Ah-Fong on the diverse signals that occur between plants and their oomycete pathogens. The authors start by describing the life- and infectivity-cycles of disease-causing oomycetes, including the familiar plant pathogens, “aggressive” Pythium and “stealthy” Phytophthora. They then cover the exchanges between host and microbe at the barrier between them. These include signals that reveal and recognize the presence of the pathogen, opportunities for pathogen entry, plant defenses and pathogen counter-defenses, nutrient exchange, and the “exit strategy” through which spores escape to start the cycle anew. The authors also expand on the formation and function of the specialized interface of the haustoria. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.18.00979
Molecular digitization of a botanical garden: high-depth whole-genome sequencing of 689 vascular plant species from the Ruili Botanical Garden
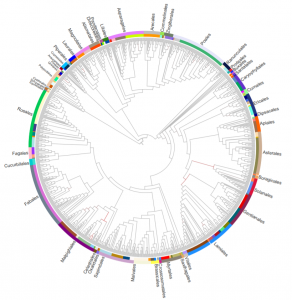 Recent advances in genome sequencing have increased our understanding of plant evolution and provided insights into factors affecting agriculturally important traits (e.g., yield, stress tolerance). There is an impetus to increase the quantity and quality of genomes publically available with around 350 of the 391,000 vascular plant species having received sequencing. Here, Liu et al. produce genomic data for 689 vascular plant species from Ruili Botanical Garden in China. Based on 78 highly conserved chloroplast genes, they reconstruct the phylogeny of vascular plants. Potential uses of their genomic dataset include advancing species identification, genome evolution and genome assembly. The project’s scale paves the way for global scale genome schemes such as the 10,000 Plant Genomes Project and the Earth BioGenome Project (Summary by Alex Bowles) Giga Science 10.1093/gigascience/giz007
Recent advances in genome sequencing have increased our understanding of plant evolution and provided insights into factors affecting agriculturally important traits (e.g., yield, stress tolerance). There is an impetus to increase the quantity and quality of genomes publically available with around 350 of the 391,000 vascular plant species having received sequencing. Here, Liu et al. produce genomic data for 689 vascular plant species from Ruili Botanical Garden in China. Based on 78 highly conserved chloroplast genes, they reconstruct the phylogeny of vascular plants. Potential uses of their genomic dataset include advancing species identification, genome evolution and genome assembly. The project’s scale paves the way for global scale genome schemes such as the 10,000 Plant Genomes Project and the Earth BioGenome Project (Summary by Alex Bowles) Giga Science 10.1093/gigascience/giz007
Modeling crop yield changes due to increased photosynthetic capabilities ($)
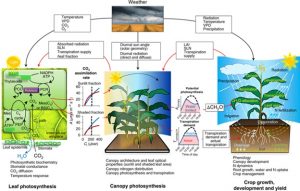 With the need to feed the growing population and the threat of global climate change, there is an imminent need to increase crop yields. One commonly accepted method of accomplishing this is by enhancing the photosynthetic capability of major crop plants, which may result in an increased yield. A recent study from Wu et al. attempted to quantify the impact that modifying photosynthetic capabilities would have in major crop plants. They accomplished this by developing a cross-scale model to connect a variety of disparate factors including interactions with crop growth, the dynamics of plant development, and environmental conditions. The authors found high variance in their models, particularly caused by the stomatal conductance of the crop and availability of water. They noted that choosing which photosynthetic components should be modified will depend greatly on the type of crop, rather than taking an all-or-nothing approach. Taken together with climate change models, this model could allow for an effective evaluation of how engineering photosynthesis will affect major food producing crops in an ever-changing climate. (Summary by Nick Segerson) Nature Plants 10.1038/s41477-019-0398-8
With the need to feed the growing population and the threat of global climate change, there is an imminent need to increase crop yields. One commonly accepted method of accomplishing this is by enhancing the photosynthetic capability of major crop plants, which may result in an increased yield. A recent study from Wu et al. attempted to quantify the impact that modifying photosynthetic capabilities would have in major crop plants. They accomplished this by developing a cross-scale model to connect a variety of disparate factors including interactions with crop growth, the dynamics of plant development, and environmental conditions. The authors found high variance in their models, particularly caused by the stomatal conductance of the crop and availability of water. They noted that choosing which photosynthetic components should be modified will depend greatly on the type of crop, rather than taking an all-or-nothing approach. Taken together with climate change models, this model could allow for an effective evaluation of how engineering photosynthesis will affect major food producing crops in an ever-changing climate. (Summary by Nick Segerson) Nature Plants 10.1038/s41477-019-0398-8
A jasmonate signaling network activates root stem cells and promotes regeneration ($)
 Plants possess plasticity for regeneration of organs after damage by physical, biotic or abiotic stress. The mitotically less-active organizer cells, quiescent center (QC) and surrounding initials form the stem cell niche, which is known to play a very important role in activation of the regeneration process in animals as well as in plants. In the Arabidopsis regeneration activation pathway, the RETINOBLASTOMA RELATED (RBR)-SCARECROW (SCR)-SHORT ROOT (SHR) protein network regulates cell division in root stem cells. Stress hormones such as JA, abscisic acid and ethylene can regulate cell division and may take part in the regeneration process. Here, Zhou et al. show that the stress hormone JA has a conspicuous role in activation of regeneration by taking part in the established RBR-SCR pathway. The authors studied the JA response after wounding created by laser ablation or resection, and after biotic damage by nematode herbivory. The signaling pathway induced by JA includes AP2/ERF family transcription factors ERF115 and ERF109, which control the cell division. In summary, this study shows the important role of JA in cell regeneration pathway for roots. (Summary by Mugdha Sabale) Cell 10.1016/j.cell.2019.03.006
Plants possess plasticity for regeneration of organs after damage by physical, biotic or abiotic stress. The mitotically less-active organizer cells, quiescent center (QC) and surrounding initials form the stem cell niche, which is known to play a very important role in activation of the regeneration process in animals as well as in plants. In the Arabidopsis regeneration activation pathway, the RETINOBLASTOMA RELATED (RBR)-SCARECROW (SCR)-SHORT ROOT (SHR) protein network regulates cell division in root stem cells. Stress hormones such as JA, abscisic acid and ethylene can regulate cell division and may take part in the regeneration process. Here, Zhou et al. show that the stress hormone JA has a conspicuous role in activation of regeneration by taking part in the established RBR-SCR pathway. The authors studied the JA response after wounding created by laser ablation or resection, and after biotic damage by nematode herbivory. The signaling pathway induced by JA includes AP2/ERF family transcription factors ERF115 and ERF109, which control the cell division. In summary, this study shows the important role of JA in cell regeneration pathway for roots. (Summary by Mugdha Sabale) Cell 10.1016/j.cell.2019.03.006
Network paths for nitrogen response in Arabidopsis
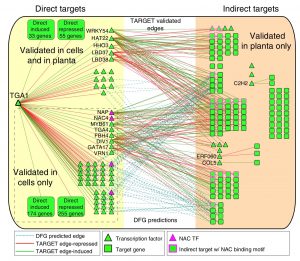 A goal of systems biology is to study how transcription factors participate in gene regulatory networks (GRN) underlying biological processes. In a recent report, Brooks et al. used network science to uncover how transcription factors mediate the early response to nitrogen (N). Authors identified the direct targets of 33 transcription factors using an in vivo approach called TARGET (Transient Assay Reporting Genome-wide Effects of Transcription factors), which detects TF-induced changes in gene expression. TARGET-data provides an excellent complementation to previously reported yeast one-hybrid (Y1H) and DNA affinity purification sequencing (DAP-seq) datasets. With all these tools together, a better understanding of TF-DNA binding can be achieved. Using Network Walking (a method for identifying the path of a TF to other downstream TFs and indirect targets), the authors reconstruct the order of TF-target interactions underlying the gene regulatory networks for TGA1 and CRF4, two important regulators of N response. There is still a lot of data to explore since 24 of the tested TF have an unknown role in N signaling. The generation of this network could have a direct impact on understanding and manipulation of GRN underlying nitrogen use in plants. (Summary by Humberto Herrera-Umbaldo) Nature Comms. 10.1038/s41467-019-09522-1
A goal of systems biology is to study how transcription factors participate in gene regulatory networks (GRN) underlying biological processes. In a recent report, Brooks et al. used network science to uncover how transcription factors mediate the early response to nitrogen (N). Authors identified the direct targets of 33 transcription factors using an in vivo approach called TARGET (Transient Assay Reporting Genome-wide Effects of Transcription factors), which detects TF-induced changes in gene expression. TARGET-data provides an excellent complementation to previously reported yeast one-hybrid (Y1H) and DNA affinity purification sequencing (DAP-seq) datasets. With all these tools together, a better understanding of TF-DNA binding can be achieved. Using Network Walking (a method for identifying the path of a TF to other downstream TFs and indirect targets), the authors reconstruct the order of TF-target interactions underlying the gene regulatory networks for TGA1 and CRF4, two important regulators of N response. There is still a lot of data to explore since 24 of the tested TF have an unknown role in N signaling. The generation of this network could have a direct impact on understanding and manipulation of GRN underlying nitrogen use in plants. (Summary by Humberto Herrera-Umbaldo) Nature Comms. 10.1038/s41467-019-09522-1
Genetic compensation mechanisms for maintaining plant stem cell robustness ($)
 In shoot meristems, the CLAVATA (CLV)-WUSCHEL(WUS) signaling module contributes to the continuous stem cell proliferation for plant development. In Arabidopsis, core CLV-WUS signaling includes the peptide ligand CLV3 and its receptor protein kinase CLV1 together with the transcription factor WUS. These proteins form a deeply conserved negative feedback loop to regulate the stem cell homeostasis. Since both the CLV3/ CLE ligands and their receptors belong to large gene families, whether they have gene compensation and how it occurs are critical questions that plant scientists would like to understand. To learn more about this transcriptional compensation, Rodriguez-Leal et al. systematically analyzed the CLV3 paralogs and found that compensation among ligand and receptor paralogs is critical for stem cell homeostasis, but that diverse genetic mechanisms buffer conserved developmental programs. Both active and passive gene compensation effects were observed in dicot and monocot plant species. This study provides good examples to explain genetic compensation mechanisms among plants and contributes to plant bioengineering for precise control of plant stem cell proliferation. (Summary by Jin Liao) Nature Genetics 10.1038/s41588-019-0389-8
In shoot meristems, the CLAVATA (CLV)-WUSCHEL(WUS) signaling module contributes to the continuous stem cell proliferation for plant development. In Arabidopsis, core CLV-WUS signaling includes the peptide ligand CLV3 and its receptor protein kinase CLV1 together with the transcription factor WUS. These proteins form a deeply conserved negative feedback loop to regulate the stem cell homeostasis. Since both the CLV3/ CLE ligands and their receptors belong to large gene families, whether they have gene compensation and how it occurs are critical questions that plant scientists would like to understand. To learn more about this transcriptional compensation, Rodriguez-Leal et al. systematically analyzed the CLV3 paralogs and found that compensation among ligand and receptor paralogs is critical for stem cell homeostasis, but that diverse genetic mechanisms buffer conserved developmental programs. Both active and passive gene compensation effects were observed in dicot and monocot plant species. This study provides good examples to explain genetic compensation mechanisms among plants and contributes to plant bioengineering for precise control of plant stem cell proliferation. (Summary by Jin Liao) Nature Genetics 10.1038/s41588-019-0389-8
Transposable elements drive rapid phenotypic variation in Capsella rubella
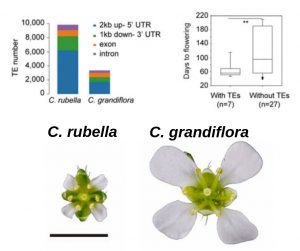 How can a species with limited genetic variation rapidly adapt to new environments? This phenomenon is known as the genetic paradox of invasion. Here, Niu et al. studied the role of Transposable Elements (TEs) in Capsella rubella, focusing on flowering-time variation. By analyzing population genomics and comparing with its outcrossing sister C. grandiflora, TEs were found “highly enriched and polymorphic in promoters and downstream regions of C. rubella genes”, mostly in intergenic regions. Sequencing revealed frequent TE insertions in the 3’UTR of Flowering Locus C (FLC), contributing to a decrease in FLC expression and early flowering. The authors postulated CrFLC expression decreases due to altered mRNA stability. 3’RACE and GUS-reporter assays showed alternative polyadenylation caused by the insertions, which leads to faster degradation of CrFLC and explained its decrease. This study suggests TE regulations are potential agents of rapid adaptive diversification, conferring phenotypic advantages in species affected by genetic bottlenecks. (Summary by Ana Valladares). Proc. Natl. Acad. Sci. USA 10.1073/pnas.1811498116
How can a species with limited genetic variation rapidly adapt to new environments? This phenomenon is known as the genetic paradox of invasion. Here, Niu et al. studied the role of Transposable Elements (TEs) in Capsella rubella, focusing on flowering-time variation. By analyzing population genomics and comparing with its outcrossing sister C. grandiflora, TEs were found “highly enriched and polymorphic in promoters and downstream regions of C. rubella genes”, mostly in intergenic regions. Sequencing revealed frequent TE insertions in the 3’UTR of Flowering Locus C (FLC), contributing to a decrease in FLC expression and early flowering. The authors postulated CrFLC expression decreases due to altered mRNA stability. 3’RACE and GUS-reporter assays showed alternative polyadenylation caused by the insertions, which leads to faster degradation of CrFLC and explained its decrease. This study suggests TE regulations are potential agents of rapid adaptive diversification, conferring phenotypic advantages in species affected by genetic bottlenecks. (Summary by Ana Valladares). Proc. Natl. Acad. Sci. USA 10.1073/pnas.1811498116
Pollinators and herbivores boost the evolution of floral traits ($)
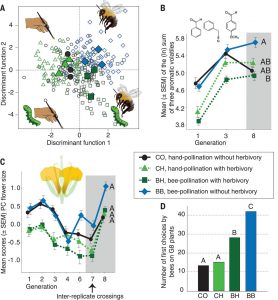 The characteristics of flowers, including morphology, color, nectar and fragrance, are closely related to their pollination methods. To attract pollinators, plants often produce large, colorful, or fragrant flowers. Increasing evidence has indicated that herbivorous feeding activity also affects flower evolution, but few studies look at the effects of both pollination and insect gnawing on flower evolution. Ramos and Schiestl used an experimental evolution strategy to observe floral characteristics in Brassica rapa in response to pollinating and/or herbivorous insects. After only six generations, there were significant differences among different experimental groups. The flowers of plants pollinated by bumble bee were more attractive and fragrant than those pollinated by artificial pollination, but only in the absence of herbivorous insects (Pieris brassicae caterpillars). In the presence of leaf-eating insects, the flowers developed self-pollinating characteristics (e.g., less spatial separation between sexual organs) and greater self-compatibility. The authors suggest that in the presence of herbivores, resources are directed towards defense rather than floral attractiveness. The study reveals that different environmental factors have non-additive effects on flower morphology, (Summary by Nanxun Qin). Science 10.1126/science.aav6962
The characteristics of flowers, including morphology, color, nectar and fragrance, are closely related to their pollination methods. To attract pollinators, plants often produce large, colorful, or fragrant flowers. Increasing evidence has indicated that herbivorous feeding activity also affects flower evolution, but few studies look at the effects of both pollination and insect gnawing on flower evolution. Ramos and Schiestl used an experimental evolution strategy to observe floral characteristics in Brassica rapa in response to pollinating and/or herbivorous insects. After only six generations, there were significant differences among different experimental groups. The flowers of plants pollinated by bumble bee were more attractive and fragrant than those pollinated by artificial pollination, but only in the absence of herbivorous insects (Pieris brassicae caterpillars). In the presence of leaf-eating insects, the flowers developed self-pollinating characteristics (e.g., less spatial separation between sexual organs) and greater self-compatibility. The authors suggest that in the presence of herbivores, resources are directed towards defense rather than floral attractiveness. The study reveals that different environmental factors have non-additive effects on flower morphology, (Summary by Nanxun Qin). Science 10.1126/science.aav6962
Meta-analysis reveals pollinator functional diversity and abundance enhance crop pollination and yield
 Insects provide a valuable service to the agricultural industry through pollination, which increases both the quality and harvest volume for many important food crops, however little is known regarding the role of insect functional trait differences in promoting crop pollination. Woodcock et al. tested two contrasting hypotheses that propose mechanisms through which pollinator community structure and functional traits influence pollination: the mass ratio hypothesis, which posits that the traits of the most abundant insect species will predict pollination success in a community, and the complementarity hypothesis, which posits that communities with more diverse functional traits will have greater pollination success. They found support for the complementarity hypothesis via correlations between functional divergence in effect traits and crop yield after accounting for any effects of overall pollinator abundance. No evidence supporting the mass ratio hypothesis was found. This study highlights the potential value that promoting and maintaining functionally diverse insect communities could have for the agricultural industry and the importance of further studying the relationships between pollinator functional diversity and plant fitness. (Summary by Rebecca Hayes) Nature Comms. 10.1038/s41467-019-09393-6
Insects provide a valuable service to the agricultural industry through pollination, which increases both the quality and harvest volume for many important food crops, however little is known regarding the role of insect functional trait differences in promoting crop pollination. Woodcock et al. tested two contrasting hypotheses that propose mechanisms through which pollinator community structure and functional traits influence pollination: the mass ratio hypothesis, which posits that the traits of the most abundant insect species will predict pollination success in a community, and the complementarity hypothesis, which posits that communities with more diverse functional traits will have greater pollination success. They found support for the complementarity hypothesis via correlations between functional divergence in effect traits and crop yield after accounting for any effects of overall pollinator abundance. No evidence supporting the mass ratio hypothesis was found. This study highlights the potential value that promoting and maintaining functionally diverse insect communities could have for the agricultural industry and the importance of further studying the relationships between pollinator functional diversity and plant fitness. (Summary by Rebecca Hayes) Nature Comms. 10.1038/s41467-019-09393-6