Plant Science Research Weekly: April 15, 2022
Special issue: Sex determination and sex chromosome evolution in land plants, Philosophical Transactions B
 At some point, most biology students learn the term “dioecy” (two houses), which refers to the separation of male and female function into different individuals, a characteristic that is common in animals but less so in flowering plants. About 6% of angiosperms and 65% of gymnosperms are dioecious, including familiar plants such as asparagus, cannabis, poplar, and ginkgo. A new special issue of Philosophical Transactions B addresses all aspects of sex determination and sex chromosome evolution in land plants. The collection of papers in this issue addresses fundamental and open questions such as the evolutionary origins of dioecy in plants, which may have derived from monoecy (having distinct male and female flowers on a single plant, as in corn). Interestingly, although there are simple systems of sex determinacy by sex chromosomes (e.g., X and Y) in both animals and plants, there are also cases where sex appears to be determined by a single-gene sex switch. One intriguing article investigates whether plants, like many animals, show a sex-based difference in life span. Like many questions about sex, the answer is, “It’s complicated.” (Summary by Mary Williams @PlantTeaching) Phil. Trans. Roy. Soc. B https://royalsocietypublishing.org/toc/rstb/2022/377/1850
At some point, most biology students learn the term “dioecy” (two houses), which refers to the separation of male and female function into different individuals, a characteristic that is common in animals but less so in flowering plants. About 6% of angiosperms and 65% of gymnosperms are dioecious, including familiar plants such as asparagus, cannabis, poplar, and ginkgo. A new special issue of Philosophical Transactions B addresses all aspects of sex determination and sex chromosome evolution in land plants. The collection of papers in this issue addresses fundamental and open questions such as the evolutionary origins of dioecy in plants, which may have derived from monoecy (having distinct male and female flowers on a single plant, as in corn). Interestingly, although there are simple systems of sex determinacy by sex chromosomes (e.g., X and Y) in both animals and plants, there are also cases where sex appears to be determined by a single-gene sex switch. One intriguing article investigates whether plants, like many animals, show a sex-based difference in life span. Like many questions about sex, the answer is, “It’s complicated.” (Summary by Mary Williams @PlantTeaching) Phil. Trans. Roy. Soc. B https://royalsocietypublishing.org/toc/rstb/2022/377/1850
ATTED-II v11: A plant gene coexpression database using a sample balancing technique by subagging of principal components
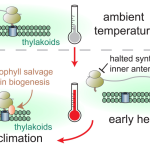
 Genes are generally expressed in concert with one another with data often represented by gene co-expression networks. Gene co-expression data may be useful in identifying genes with unknown functions and are generally utilized in exploring molecular processes occurring during normal development as well as during biological stresses. ATTED-II, is a database of plant gene coexpression data which has recently received an update with version 11. Within the database, nine plant species are represented with data originating from publicly available RNAseq and microarray studies. Using publicly available data is complicated by inherent sampling bias. ATTED-II uses principle component analysis and ensemble calculation (subagging) as methods of sample balancing to reduce difficulties in exploiting this rich source of data. The database enables users to examine coexpression networks between species which may prove beneficial for extrapolation of data from model species to plant species of interest globally. (Summary by Orla Sherwood, @orlasherwood) Plant Cell Physiol. 10.1093/pcp/pcac041)
Genes are generally expressed in concert with one another with data often represented by gene co-expression networks. Gene co-expression data may be useful in identifying genes with unknown functions and are generally utilized in exploring molecular processes occurring during normal development as well as during biological stresses. ATTED-II, is a database of plant gene coexpression data which has recently received an update with version 11. Within the database, nine plant species are represented with data originating from publicly available RNAseq and microarray studies. Using publicly available data is complicated by inherent sampling bias. ATTED-II uses principle component analysis and ensemble calculation (subagging) as methods of sample balancing to reduce difficulties in exploiting this rich source of data. The database enables users to examine coexpression networks between species which may prove beneficial for extrapolation of data from model species to plant species of interest globally. (Summary by Orla Sherwood, @orlasherwood) Plant Cell Physiol. 10.1093/pcp/pcac041)
Systems approaches reveal that ABCB and PIN proteins mediate co-dependent auxin efflux
 In plant development, advanced computer model simulations allow the understanding of the non-intuitive relations between local morphogenetic processes and global patterning. In Arabidopsis thaliana, the hormone auxin is involved in several processes of plant development. Auxin transporters play an important role in its distribution within plant tissue. The PIN-formed (PIN) and B sub-family of membrane-bound ATP-binding cassette (ABC) transporters (ABCB) are involved in local and global auxin distribution. In this study, Mellor et al. investigated the ABCB-PIN regulatory interaction in the Arabidopsis thaliana root. They used a computational model to set up five scenarios based on the ABCB-PIN function and expression pattern. In addition, they tested the distribution of DII-VENUS auxin reporter with the abcb single and double mutants. They found that the auxin distribution at the tip of the root is perturbed in abcb double mutants compared to the single mutants. They suggest that ABCBs are able to act independently of PIN. This study provides an overview of how the auxin patterns are affected by the interaction of PIN and ABCB proteins. (Summary by Andrea Gómez Felipe @andreagomezfe) Plant Cell 10.1093/plcell/koac086.
In plant development, advanced computer model simulations allow the understanding of the non-intuitive relations between local morphogenetic processes and global patterning. In Arabidopsis thaliana, the hormone auxin is involved in several processes of plant development. Auxin transporters play an important role in its distribution within plant tissue. The PIN-formed (PIN) and B sub-family of membrane-bound ATP-binding cassette (ABC) transporters (ABCB) are involved in local and global auxin distribution. In this study, Mellor et al. investigated the ABCB-PIN regulatory interaction in the Arabidopsis thaliana root. They used a computational model to set up five scenarios based on the ABCB-PIN function and expression pattern. In addition, they tested the distribution of DII-VENUS auxin reporter with the abcb single and double mutants. They found that the auxin distribution at the tip of the root is perturbed in abcb double mutants compared to the single mutants. They suggest that ABCBs are able to act independently of PIN. This study provides an overview of how the auxin patterns are affected by the interaction of PIN and ABCB proteins. (Summary by Andrea Gómez Felipe @andreagomezfe) Plant Cell 10.1093/plcell/koac086.
KAI2 regulates seedling development by mediating light-induced remodeling of auxin transport
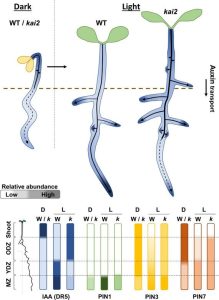 Seed germination is can be described as having two phases depending on light availability, skoto- and photomorphogenesis. Skotomorphogenesis, which occurs while the seed is still buried, is characterized by the elongation of the coleoptile and the inhibition of the root system. When the seed reaches the light, photomorphogenesis starts, which includes the expansion of the cotyledons and the development of the root system. Auxin plays a crucial role during both phases, regulating both hypocotyl elongation and root formation. The karrikin insensitive2/hyposensitive to light (KAI2) Arabidopsis thaliana mutants show a deficiency in photomorphogenesis. KAI2 is closely related to DWARF14 (D-14), which acts as a receptor for strigolactone molecules. However, the role of KAI2 in seedling photomorphogenesis remains poorly understood. In this work, Hamon-Josse et al. dissect the role of KAI2 during photomorphogenesis. Briefly, the authors describe why kai2 mutant lines show a deficiency in photomorphogenesis, indicating that these lines sense light but they are unable to downregulate auxin transport from the shot apex to the roots. This change in auxin distribution leads to an enhanced development of the root system. Based on these results, the authors could suppress most kai2 seedling phenotypes by removing auxin transport-related genes (such as some genes from the PIN-FORMED family) or by the use of inhibitors of auxin synthesis or transport. In conclusion, KAI2 regulates photomorphogenesis because it promotes a large-scale remodeling of the auxin transport system that affects the abundance of PIN proteins within cells. (Summary by Eva Maria Gomez Alvarez @eva_ga96) New Phytol. 10.1111/nph.18110
Seed germination is can be described as having two phases depending on light availability, skoto- and photomorphogenesis. Skotomorphogenesis, which occurs while the seed is still buried, is characterized by the elongation of the coleoptile and the inhibition of the root system. When the seed reaches the light, photomorphogenesis starts, which includes the expansion of the cotyledons and the development of the root system. Auxin plays a crucial role during both phases, regulating both hypocotyl elongation and root formation. The karrikin insensitive2/hyposensitive to light (KAI2) Arabidopsis thaliana mutants show a deficiency in photomorphogenesis. KAI2 is closely related to DWARF14 (D-14), which acts as a receptor for strigolactone molecules. However, the role of KAI2 in seedling photomorphogenesis remains poorly understood. In this work, Hamon-Josse et al. dissect the role of KAI2 during photomorphogenesis. Briefly, the authors describe why kai2 mutant lines show a deficiency in photomorphogenesis, indicating that these lines sense light but they are unable to downregulate auxin transport from the shot apex to the roots. This change in auxin distribution leads to an enhanced development of the root system. Based on these results, the authors could suppress most kai2 seedling phenotypes by removing auxin transport-related genes (such as some genes from the PIN-FORMED family) or by the use of inhibitors of auxin synthesis or transport. In conclusion, KAI2 regulates photomorphogenesis because it promotes a large-scale remodeling of the auxin transport system that affects the abundance of PIN proteins within cells. (Summary by Eva Maria Gomez Alvarez @eva_ga96) New Phytol. 10.1111/nph.18110
LEC1 acts as a molecular signal between the endosperm and embryo to initiate and regulate embryo maturation
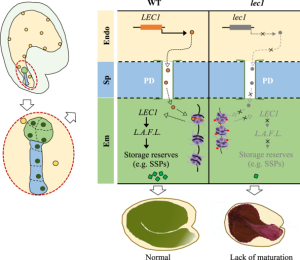 In most angiosperms, the seed is an outcome of a double fertilization process that initiates the development of the embryo and endosperm. The endosperm plays an essential role in nourishing the developing embryo by ensuring adequate maternal nutrient transfer. However, we are far from knowing the factors that coordinate the developing embryo and endosperm. LEAFY COTYLEDON 1 (LEC1) is expressed in both embryo and endosperm and plays a central role in most aspects of seed maturation. Song et al. hypothesized that the endosperm-synthesized LEC1 may act as a molecular signal between endosperm and developing embryo and eventually perform its function during embryo development. Using elaborate genetics, the authors demonstrated that endosperm-expressed LEC1 is sufficient for seed maturation while exclusive embryonic LEC1 expression led to embryo developmental arrest. Furthermore, the authors revealed that the LEC1 protein is synthesized in the endosperm and trafficked to the embryo via the suspensor to perform its function. The authors speculated that the chromatin and cellular conditions in the endosperm favor the synthesis of LEC1 shortly after fertilization. Then, it enters the embryo and activates the transcription of seed maturation-specific genes, including itself. (Summary by Subodh Verma @subodhshanky) Nature Comms. 10.1038/s41467-021-24234-1
In most angiosperms, the seed is an outcome of a double fertilization process that initiates the development of the embryo and endosperm. The endosperm plays an essential role in nourishing the developing embryo by ensuring adequate maternal nutrient transfer. However, we are far from knowing the factors that coordinate the developing embryo and endosperm. LEAFY COTYLEDON 1 (LEC1) is expressed in both embryo and endosperm and plays a central role in most aspects of seed maturation. Song et al. hypothesized that the endosperm-synthesized LEC1 may act as a molecular signal between endosperm and developing embryo and eventually perform its function during embryo development. Using elaborate genetics, the authors demonstrated that endosperm-expressed LEC1 is sufficient for seed maturation while exclusive embryonic LEC1 expression led to embryo developmental arrest. Furthermore, the authors revealed that the LEC1 protein is synthesized in the endosperm and trafficked to the embryo via the suspensor to perform its function. The authors speculated that the chromatin and cellular conditions in the endosperm favor the synthesis of LEC1 shortly after fertilization. Then, it enters the embryo and activates the transcription of seed maturation-specific genes, including itself. (Summary by Subodh Verma @subodhshanky) Nature Comms. 10.1038/s41467-021-24234-1
Nitrate transporter NPF7.9 is a regulator nitrogen use efficiency and stress-induced nitrate allocation to roots in rice
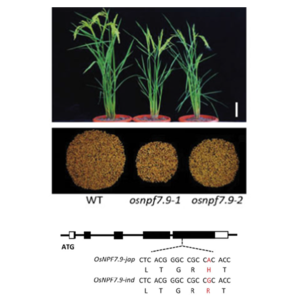 In addition to its role as a nutrient and a developmental signal, nitrate regulates stress responses in plants. By homology to nitrate transporters previously identified in Arabidopsis as contributing to stress induced-nitrate allocation to roots (SINAR) and nitrogen use efficiency (NUE), Guan et al. identified OsNPF7.9 as having a similar role in rice. Osnpf7.9 mutants are defective in shoot nitrate allocation and display lower shoot biomass allocation and grain yield. Furthermore, downregulated expression of OsNPF7.9 under cadmium and sodium toxicities correlates with the higher root nitrate retention feature of SINAR. This gene shows a single nucleotide polymorphism between indica and japonica rice. The nitrate transport efficient OsNPF7.9 SNP G allele was found predominantly in high NUE indica rice genotypes, while the SNP A allele was associated with japonica rice. These findings are an important addition to the accumulating evidence on nitrate use divergence of rice subspecies and could be a potent molecular marker for high NUE. (Summary by Lekshmy Sathee @lekshmysnair) Plant Physiol. 10.1093/plphys/kiac044
In addition to its role as a nutrient and a developmental signal, nitrate regulates stress responses in plants. By homology to nitrate transporters previously identified in Arabidopsis as contributing to stress induced-nitrate allocation to roots (SINAR) and nitrogen use efficiency (NUE), Guan et al. identified OsNPF7.9 as having a similar role in rice. Osnpf7.9 mutants are defective in shoot nitrate allocation and display lower shoot biomass allocation and grain yield. Furthermore, downregulated expression of OsNPF7.9 under cadmium and sodium toxicities correlates with the higher root nitrate retention feature of SINAR. This gene shows a single nucleotide polymorphism between indica and japonica rice. The nitrate transport efficient OsNPF7.9 SNP G allele was found predominantly in high NUE indica rice genotypes, while the SNP A allele was associated with japonica rice. These findings are an important addition to the accumulating evidence on nitrate use divergence of rice subspecies and could be a potent molecular marker for high NUE. (Summary by Lekshmy Sathee @lekshmysnair) Plant Physiol. 10.1093/plphys/kiac044
Always prepared: priming of the defense response by trichomes
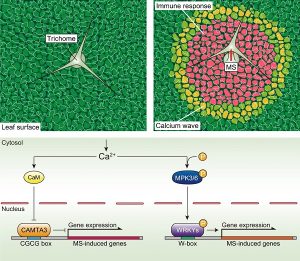 Rain contributes to plant disease. Rain can contain high concentrations of pathogens (like Pseudomonas, Xanthomonas, Alternaria, Fusarium sp., etc.), and high humidity associated with rain antagonizes stomatal closure, leaving open entry points for some pathogens. Matsumura and colleagues have demonstrated a novel function of Arabidopsis mechanosensory cells present in the leaves, the trichomes. The authors showed that raindrops can create a mechanosensation sensed by the trichomes and perceived as an emergency signal, which triggers an immune response. Upon mechanostimulation by rain, trichomes initiate an intercellular Ca2+ wave, resulting in (a) the activation of CALMODULIN-BINDING TRANSCRIPTION ACTIVATOR 3 (CAMTA3) and CAMTA3-dependent gene expression, and (b) phosphorylation of MITOGEN -ACTIVATED PROTEIN KINASE3 (MAPK3) and MAPK6 resulting in the phosphorylation of WRKY DNA-binding protein and WRKY-dependent gene expression; these are canonoical defense pathways. This work demonstrates a central role played by trichomes in mechanosensation and activation of plant defense responses for priming against an impending pathogen attack. (Summary by Sibaji K Sanyal @SibajiSanyal) Nature Comms. 10.1038/s41467-022-28813-8
Rain contributes to plant disease. Rain can contain high concentrations of pathogens (like Pseudomonas, Xanthomonas, Alternaria, Fusarium sp., etc.), and high humidity associated with rain antagonizes stomatal closure, leaving open entry points for some pathogens. Matsumura and colleagues have demonstrated a novel function of Arabidopsis mechanosensory cells present in the leaves, the trichomes. The authors showed that raindrops can create a mechanosensation sensed by the trichomes and perceived as an emergency signal, which triggers an immune response. Upon mechanostimulation by rain, trichomes initiate an intercellular Ca2+ wave, resulting in (a) the activation of CALMODULIN-BINDING TRANSCRIPTION ACTIVATOR 3 (CAMTA3) and CAMTA3-dependent gene expression, and (b) phosphorylation of MITOGEN -ACTIVATED PROTEIN KINASE3 (MAPK3) and MAPK6 resulting in the phosphorylation of WRKY DNA-binding protein and WRKY-dependent gene expression; these are canonoical defense pathways. This work demonstrates a central role played by trichomes in mechanosensation and activation of plant defense responses for priming against an impending pathogen attack. (Summary by Sibaji K Sanyal @SibajiSanyal) Nature Comms. 10.1038/s41467-022-28813-8
A mitochondrial RNA processing protein mediates plant immunity to a broad spectrum of pathogens by modulating the mitochondrial oxidative burst
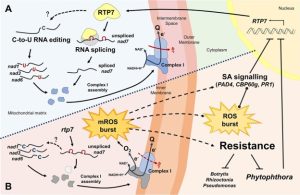 Phytophthora are a group of oomycete pathogens that infect a wide range of plants and cause disease in many important crop species. Large scale mutant screens have previously been used to identify Arabidopsis thaliana mutants with enhanced resistance to P. parasitica. Here, Yang and colleagues investigate the mechanisms underlying resistance in one such mutant, rtp7. The researchers show that RTP7, a nuclear encoded protein, localizes to mitochondria. There it participates in RNA splicing of nad7 transcripts, a mitochondrial gene encoding one of the subunits of mitochondrial respiratory chain Complex I. The group examined mitochondrial function in rtp7 plants and found decreased Complex I activity as well as increased expression of the alternative oxidase family genes, suggesting elevated production of mitochondrial reactive oxygen species (ROS). The researchers then used chemical ROS scavengers as well as plants deficient in the plasma membrane NADPH oxidases RBOHD and RBOHF to show that the increased resistance in rtp7 plants is due to mitochondrial ROS, and may be independent of the plasma membrane localized ROS bursts. Finally, the group used phylogenetic techniques to show that RTP7 is highly conserved and confirmed that silencing of rtp7 in Nicotiana benthamiana and Solanum lycopersicum also results in enhanced resistance to P. infestans. (Summary by Rory Burke @Rorby95) Plant Cell 10.1093/plcell/koac082
Phytophthora are a group of oomycete pathogens that infect a wide range of plants and cause disease in many important crop species. Large scale mutant screens have previously been used to identify Arabidopsis thaliana mutants with enhanced resistance to P. parasitica. Here, Yang and colleagues investigate the mechanisms underlying resistance in one such mutant, rtp7. The researchers show that RTP7, a nuclear encoded protein, localizes to mitochondria. There it participates in RNA splicing of nad7 transcripts, a mitochondrial gene encoding one of the subunits of mitochondrial respiratory chain Complex I. The group examined mitochondrial function in rtp7 plants and found decreased Complex I activity as well as increased expression of the alternative oxidase family genes, suggesting elevated production of mitochondrial reactive oxygen species (ROS). The researchers then used chemical ROS scavengers as well as plants deficient in the plasma membrane NADPH oxidases RBOHD and RBOHF to show that the increased resistance in rtp7 plants is due to mitochondrial ROS, and may be independent of the plasma membrane localized ROS bursts. Finally, the group used phylogenetic techniques to show that RTP7 is highly conserved and confirmed that silencing of rtp7 in Nicotiana benthamiana and Solanum lycopersicum also results in enhanced resistance to P. infestans. (Summary by Rory Burke @Rorby95) Plant Cell 10.1093/plcell/koac082
Seed microbiota revealed by a large-scale meta-analysis including 50 plant species
 The study of seed microbiota is rapidly getting attention among plant scientists, given its role in modulating seed germination and plant fitness. Here, Simonin and colleagues synthesize the information from 63 studies and provide a thorough overview of this topic’s current state of the art. The authors found that only 50 plants species have at least one study of their seed microbiota. Still, microbial communities associated with seeds were highly diverse, exhibiting considerable variation between and within species. Unlike in the microbiota of other plant organs, bacterial and fungi clades were equally prevalent in seed microbiota. Four bacterial phyla (Proteobacteria, Actinobacteria, Firmicutes and Bacteroidetes) and three fungal classes (Dothideomycetes, Sordariomycetes and Tremellomycetes) were consistently found in great proportion and across various plant species. Thus, they are considered to form the seeds’ core microbiota. While the characterization of the seed microbiota is still far from being complete, this research shows emerging patterns and traces an exciting roadmap for future studies in the area. Thus, it is obligate reading for fans of seed-associated microorganisms. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) New Phytol. 10.1111/nph.18037
The study of seed microbiota is rapidly getting attention among plant scientists, given its role in modulating seed germination and plant fitness. Here, Simonin and colleagues synthesize the information from 63 studies and provide a thorough overview of this topic’s current state of the art. The authors found that only 50 plants species have at least one study of their seed microbiota. Still, microbial communities associated with seeds were highly diverse, exhibiting considerable variation between and within species. Unlike in the microbiota of other plant organs, bacterial and fungi clades were equally prevalent in seed microbiota. Four bacterial phyla (Proteobacteria, Actinobacteria, Firmicutes and Bacteroidetes) and three fungal classes (Dothideomycetes, Sordariomycetes and Tremellomycetes) were consistently found in great proportion and across various plant species. Thus, they are considered to form the seeds’ core microbiota. While the characterization of the seed microbiota is still far from being complete, this research shows emerging patterns and traces an exciting roadmap for future studies in the area. Thus, it is obligate reading for fans of seed-associated microorganisms. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) New Phytol. 10.1111/nph.18037
A time to look for alternative nutrition sources
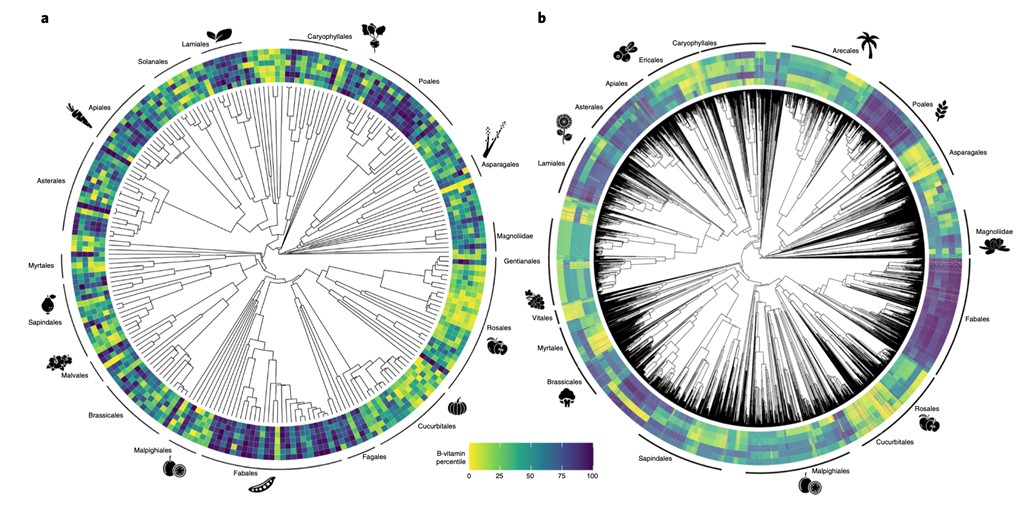 Recently, there has been a huge impetus to develop plants that can perform well under stress conditions and produce high yields so that an ever-increasing human population can be fed. Another point, less discussed but equally important, is the nutritional value that these crops can provide. Cantwell-Jones and colleagues have taken a very interesting phylogenomics investigation into the diversity of edible plants that are good sources of several types of vitamin B (thiamine, riboflavin, niacin, pantothenic acid, pyridoxine, and folate). The authors generated an edible plant phylogeny, mapped onto it species with known vitamin B levels, and predicted species that are anticipated to produce high levels of vitamin B. Although the identified species need experimental validation, theoretically they provide us with alternative sources beyond those already known. Their study noted that a significantly high number (about 42%) of the identified species are not conserved ex-situ in seed banks. This is alarming, as 15% of these species are threatened in their natural environment. Several countries can benefit by employing strategies for the conservation of these plants as they have the potential to serve as alternate vitamin B sources. Perhaps, as more countries are allowing gene editing in plants to create ‘future ready’ crops, we should also work diligently to save the ones with known benefits. (Summary by Sibaji K Sanyal @SibajiSanyal) Nature Plants 10.1038/s41477-022-01100-6
Recently, there has been a huge impetus to develop plants that can perform well under stress conditions and produce high yields so that an ever-increasing human population can be fed. Another point, less discussed but equally important, is the nutritional value that these crops can provide. Cantwell-Jones and colleagues have taken a very interesting phylogenomics investigation into the diversity of edible plants that are good sources of several types of vitamin B (thiamine, riboflavin, niacin, pantothenic acid, pyridoxine, and folate). The authors generated an edible plant phylogeny, mapped onto it species with known vitamin B levels, and predicted species that are anticipated to produce high levels of vitamin B. Although the identified species need experimental validation, theoretically they provide us with alternative sources beyond those already known. Their study noted that a significantly high number (about 42%) of the identified species are not conserved ex-situ in seed banks. This is alarming, as 15% of these species are threatened in their natural environment. Several countries can benefit by employing strategies for the conservation of these plants as they have the potential to serve as alternate vitamin B sources. Perhaps, as more countries are allowing gene editing in plants to create ‘future ready’ crops, we should also work diligently to save the ones with known benefits. (Summary by Sibaji K Sanyal @SibajiSanyal) Nature Plants 10.1038/s41477-022-01100-6