Plant Science Research Weekly: April 11, 2025
Review: Celebrating 150 years of Arabidopsis genetics
 The first known report of an Arabidopsis thaliana mutant appeared approximately 150 years ago (1873). In the intervening years, Arabidopsis has become an essential model for plant genetic research, driving groundbreaking discoveries across multiple disciplines. In a recent review, Yaschenko et al. provide a broad and succinct examination of how Arabidopsis has contributed more than its weight in scientific advancement. Its small genome, short life cycle, and ease of genetic manipulation have made it an ideal model organism, leading to major advancements in understanding plant growth, development, disease resistance, and stress tolerance. The ability to translate these findings to economically important crops has kept it as an essential tool in addressing agricultural challenges facing the world. Beyond plant biology, Arabidopsis has contributed greatly to medical research by identifying orthologs linked to human diseases, aiding in the discovery of genetic markers and biological processes relevant to potential treatments. The wealth of genomic data from Arabidopsis has also advanced biotechnology, leading to the development of tools like inducible expression systems, optogenetics, and protein interaction regulators. As research continues to evolve, Arabidopsis remains an ever-important model organism, offering critical insights into fundamental biological mechanisms that have applications beyond plant biology. (Summary by Xavier Ozowara [email protected]). Plant Cell 10.1093/plcell/koae065
The first known report of an Arabidopsis thaliana mutant appeared approximately 150 years ago (1873). In the intervening years, Arabidopsis has become an essential model for plant genetic research, driving groundbreaking discoveries across multiple disciplines. In a recent review, Yaschenko et al. provide a broad and succinct examination of how Arabidopsis has contributed more than its weight in scientific advancement. Its small genome, short life cycle, and ease of genetic manipulation have made it an ideal model organism, leading to major advancements in understanding plant growth, development, disease resistance, and stress tolerance. The ability to translate these findings to economically important crops has kept it as an essential tool in addressing agricultural challenges facing the world. Beyond plant biology, Arabidopsis has contributed greatly to medical research by identifying orthologs linked to human diseases, aiding in the discovery of genetic markers and biological processes relevant to potential treatments. The wealth of genomic data from Arabidopsis has also advanced biotechnology, leading to the development of tools like inducible expression systems, optogenetics, and protein interaction regulators. As research continues to evolve, Arabidopsis remains an ever-important model organism, offering critical insights into fundamental biological mechanisms that have applications beyond plant biology. (Summary by Xavier Ozowara [email protected]). Plant Cell 10.1093/plcell/koae065
Review. Genetic switchboards: Rewiring plant traits with synthetic circuits
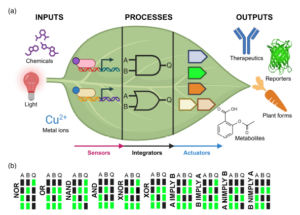 The expression of a transgene in plants can impose a significant stress, sometimes referred to as metabolic burden. Synthetic gene circuits offer a precise approach to engineering plant traits by regulating gene expression through programmable operations. This review by Lloyd et al. examines the core principles and components of these circuits, including sensors, integrators, and actuators. While natural gene regulatory networks have evolved for survival and been further modified by selective breeding, synthetic circuits provide targeted control over gene expression. These circuits function through logical operations (e.g., AND, OR, NOR gates) and require orthogonality, a principle that relies on genetic parts designed to interact strongly with each other while minimizing unintended interactions with other cellular components. The authors outline synthetic gene circuit architecture: sensors detect molecular or environmental inputs via inducible promoters, though stability challenges must be addressed. Integrators process signals using engineered promoters, recombinases, or CRISPR repressors, enabling logic-based regulation. Actuators execute the response, modifying cell function, such as controlling endogenous genes or influencing metabolic pathways. Bacterial allosteric transcription factors (aTFs) offer a promising means of combining sensing of specific metabolites and regulated gene expression but require further optimization to function efficiently in plant systems. A major challenge in plant synthetic biology is the long development time compared to bacteria, where rapid design-build-test-learn (DBTL) cycles enable faster refinement. Transient expression systems can accelerate testing before stable transformation. Other challenges include inefficient gene targeting, lack of standardized DNA delivery methods, and whole-plant regeneration constraints. Advances in computational modeling, high-throughput screening, and targeted transgene integration will be critical for progress. Overcoming these obstacles will unlock new plant traits, improve crop resilience, and enhance fundamental plant research. (Summary by Elisa De Meo) Plant J. 10.1111/tpj.70090.
The expression of a transgene in plants can impose a significant stress, sometimes referred to as metabolic burden. Synthetic gene circuits offer a precise approach to engineering plant traits by regulating gene expression through programmable operations. This review by Lloyd et al. examines the core principles and components of these circuits, including sensors, integrators, and actuators. While natural gene regulatory networks have evolved for survival and been further modified by selective breeding, synthetic circuits provide targeted control over gene expression. These circuits function through logical operations (e.g., AND, OR, NOR gates) and require orthogonality, a principle that relies on genetic parts designed to interact strongly with each other while minimizing unintended interactions with other cellular components. The authors outline synthetic gene circuit architecture: sensors detect molecular or environmental inputs via inducible promoters, though stability challenges must be addressed. Integrators process signals using engineered promoters, recombinases, or CRISPR repressors, enabling logic-based regulation. Actuators execute the response, modifying cell function, such as controlling endogenous genes or influencing metabolic pathways. Bacterial allosteric transcription factors (aTFs) offer a promising means of combining sensing of specific metabolites and regulated gene expression but require further optimization to function efficiently in plant systems. A major challenge in plant synthetic biology is the long development time compared to bacteria, where rapid design-build-test-learn (DBTL) cycles enable faster refinement. Transient expression systems can accelerate testing before stable transformation. Other challenges include inefficient gene targeting, lack of standardized DNA delivery methods, and whole-plant regeneration constraints. Advances in computational modeling, high-throughput screening, and targeted transgene integration will be critical for progress. Overcoming these obstacles will unlock new plant traits, improve crop resilience, and enhance fundamental plant research. (Summary by Elisa De Meo) Plant J. 10.1111/tpj.70090.
Cells are larger than life when ExPOSEd
 Cells are as small as life gets, but can be much larger than they appear if given room for expansion. This is possible with expansion microscopy, a technique that enables three dimensional cell imaging by physically expanding cellular components for visualization. Although expansion microscopy has been used in several eukaryotic systems, it is only recently being extended to plant cells, which present challenges due to their cell walls. To address this problem, Cox et al. introduce ExPOSE, an expansion microscopy technique that has been optimized for plant protoplasts – cells without cell walls. The cell wall of maize and Arabidopsis leaf cells were enzymatically digested to isolate protoplasts which were then fixed, treated with a protein-binding anchor, and embedded in a swellable hydrogel overnight. This process resulted in an average physical expansion of more than 10-fold. With the cell physically zoomed in, nothing stays hidden. ExPOSE allows high-resolution visualization of cell components such as protein localization within mitochondrial matrices which are normally invisible in unexpanded cells. The authors also used ExPOSE to observe DNA architecture, detect individual mRNA foci, resolve the spatial resolution of tightly packed proteins, and capture subtle co-localization changes – all using a standard confocal microscope. Perhaps one of ExPOSE’s standout advantages is its application in studying biomolecular condensates, a use not previously reported with expansion microscopy. If single cell study is the target, then there’s room for expansion with ExPOSE. (Summary by Irene I. Ikiriko @ireneikiriko) Plant J. 10.1111/tpj.70049
Cells are as small as life gets, but can be much larger than they appear if given room for expansion. This is possible with expansion microscopy, a technique that enables three dimensional cell imaging by physically expanding cellular components for visualization. Although expansion microscopy has been used in several eukaryotic systems, it is only recently being extended to plant cells, which present challenges due to their cell walls. To address this problem, Cox et al. introduce ExPOSE, an expansion microscopy technique that has been optimized for plant protoplasts – cells without cell walls. The cell wall of maize and Arabidopsis leaf cells were enzymatically digested to isolate protoplasts which were then fixed, treated with a protein-binding anchor, and embedded in a swellable hydrogel overnight. This process resulted in an average physical expansion of more than 10-fold. With the cell physically zoomed in, nothing stays hidden. ExPOSE allows high-resolution visualization of cell components such as protein localization within mitochondrial matrices which are normally invisible in unexpanded cells. The authors also used ExPOSE to observe DNA architecture, detect individual mRNA foci, resolve the spatial resolution of tightly packed proteins, and capture subtle co-localization changes – all using a standard confocal microscope. Perhaps one of ExPOSE’s standout advantages is its application in studying biomolecular condensates, a use not previously reported with expansion microscopy. If single cell study is the target, then there’s room for expansion with ExPOSE. (Summary by Irene I. Ikiriko @ireneikiriko) Plant J. 10.1111/tpj.70049
Expanding the resolution limits of conventional microscopy in whole plant tissues
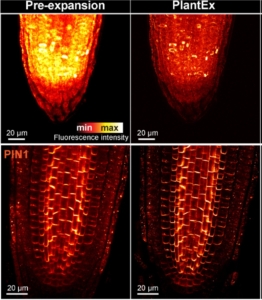 How can we precisely image plant tissues in super-resolution when approaching the optical limits of conventional microscopes? One solution lies in expansion microscopy, a technique that embeds tissue samples in an expandable hydrogel that proportionally increases the distances between structures, allowing the user to image them clearly on microscopes that would be diffraction-limited at such fine scales. While this technique has been applied to many fields of biology, and even plant protoplasts (see above), whole plant tissues present particular challenges due to their rigid and cohesive cell walls. In this paper, Gallei, Truckenbrodt and colleagues describe PlantEx, a plant-specific expansion microscopy protocol that includes a cell wall digestion step crafted to address these challenges. This process is demonstrated with Arabidopsis thaliana root tissue and the results confirmed to introduce no significant distortion to tissue architecture. PlantEx is also combined with stimulated emission depletion microscopy to further increase resolution and enable subcellular imaging. PlantEx can only be performed on fixed tissues and would require calibration for application to other plant species and tissue types, but it has transformative potential in increasing ease and accessibility of super-resolution imaging for plant biology. (Summary by Elise Krespan) Plant Cell 10.1093/plcell/koaf006
How can we precisely image plant tissues in super-resolution when approaching the optical limits of conventional microscopes? One solution lies in expansion microscopy, a technique that embeds tissue samples in an expandable hydrogel that proportionally increases the distances between structures, allowing the user to image them clearly on microscopes that would be diffraction-limited at such fine scales. While this technique has been applied to many fields of biology, and even plant protoplasts (see above), whole plant tissues present particular challenges due to their rigid and cohesive cell walls. In this paper, Gallei, Truckenbrodt and colleagues describe PlantEx, a plant-specific expansion microscopy protocol that includes a cell wall digestion step crafted to address these challenges. This process is demonstrated with Arabidopsis thaliana root tissue and the results confirmed to introduce no significant distortion to tissue architecture. PlantEx is also combined with stimulated emission depletion microscopy to further increase resolution and enable subcellular imaging. PlantEx can only be performed on fixed tissues and would require calibration for application to other plant species and tissue types, but it has transformative potential in increasing ease and accessibility of super-resolution imaging for plant biology. (Summary by Elise Krespan) Plant Cell 10.1093/plcell/koaf006
CA-nundrum: How a spontaneous mutation in carbonic anhydrase uncouples leaf δ13C, WUE and C4 photosynthesis
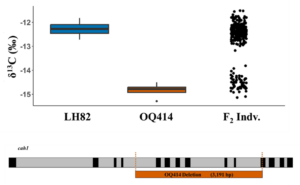 With climate change, drought is expected to happen more frequently, making supplemental irrigation increasingly necessary to sustain crop productivity. One target trait to improve climate resilience is water use efficiency (WUE), defined by the ratio of carbon assimilation to water used by the plant. However, directly measuring WUE is time consuming and low-throughput, highlighting the need for proxy traits that enable large-scale phenotyping. Leaf tissue stable carbon isotope composition (δ13Cleaf) is commonly used as a proxy for WUE because it reflects CO2 availability and carbon metabolism. In this study, Twohey et al. investigated two maize lines, OQ414 and LH82, which showed similar morphological characteristics but very different δ13Cleaf values. A biparental mapping F2 population derived from these two lines showed a simple dominance segregation pattern for δ13Cleaf. Genetic mapping identified a major QTL on chromosome 3, located within 1 LOD of the carbonic anhydrase 1 (cah1) and carbonic anhydrase 2 (cah2) loci. Complementation test confirmed that the phenotype observed was due to a mutation in cah1. Sequencing revealed that OQ414 harbors a deletion in exons 5-10 and part of exon 11 in cah1. Surprisingly, this mutation led to significantly increased carbonic anhydrase (CA) content and photosynthetic rates. The altered domain structure in OQ414 is 222 amino acids shorter than wildtype which resembles a functional splice variant. In conclusion, this study demonstrated that mutation in cah1 uncoupled the relationship between δ13C and WUE in C4 plants. The findings underscore the complexity of using δ13Cleaf as a proxy for breeding WUE in C4 species. This paper also reports, for the first time, a mutation in CA gene that does not impair, but rather enhances, CA activity. (Summary by Mae Mercado) bioRxiv https://doi.org/10.1101/2025.02.20.639358
With climate change, drought is expected to happen more frequently, making supplemental irrigation increasingly necessary to sustain crop productivity. One target trait to improve climate resilience is water use efficiency (WUE), defined by the ratio of carbon assimilation to water used by the plant. However, directly measuring WUE is time consuming and low-throughput, highlighting the need for proxy traits that enable large-scale phenotyping. Leaf tissue stable carbon isotope composition (δ13Cleaf) is commonly used as a proxy for WUE because it reflects CO2 availability and carbon metabolism. In this study, Twohey et al. investigated two maize lines, OQ414 and LH82, which showed similar morphological characteristics but very different δ13Cleaf values. A biparental mapping F2 population derived from these two lines showed a simple dominance segregation pattern for δ13Cleaf. Genetic mapping identified a major QTL on chromosome 3, located within 1 LOD of the carbonic anhydrase 1 (cah1) and carbonic anhydrase 2 (cah2) loci. Complementation test confirmed that the phenotype observed was due to a mutation in cah1. Sequencing revealed that OQ414 harbors a deletion in exons 5-10 and part of exon 11 in cah1. Surprisingly, this mutation led to significantly increased carbonic anhydrase (CA) content and photosynthetic rates. The altered domain structure in OQ414 is 222 amino acids shorter than wildtype which resembles a functional splice variant. In conclusion, this study demonstrated that mutation in cah1 uncoupled the relationship between δ13C and WUE in C4 plants. The findings underscore the complexity of using δ13Cleaf as a proxy for breeding WUE in C4 species. This paper also reports, for the first time, a mutation in CA gene that does not impair, but rather enhances, CA activity. (Summary by Mae Mercado) bioRxiv https://doi.org/10.1101/2025.02.20.639358
Lights, camera, pectin: Bringing hypocotyl elongation out of the dark
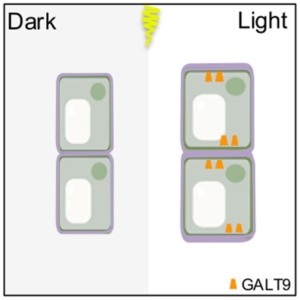 Light is a powerful signal, shaping plant development and growth. However, the cellular mechanisms that translate light signals into precise developmental responses are still being unravelled. The Arabidopsis hypocotyl (the embryonic stem, situated underneath the cotyledons in seedlings) rapidly restricts elongation upon light exposure. Zhang et al. combined molecular and mechanical techniques to uncover a succinct model for how this inhibition occurs in the dark-to-light transition. Initially, using time-lapse photography, they identified a key regulator inhibiting hypocotyl elongation: ELONGATED HYPOCOTYL 5 (HY5). Through chemical quantification and immunohistochemical analysis of the cell walls they found that, unlike in wild-type plants, there is no change in the accumulation of pectin in the hy5 mutant. Further, Raman microscopy showed that pectin is polarised to transverse walls of cells following light treatment – a pattern absent in hy5. This polarisation of pectin is due to the asymmetric localisation of GALACTOTRANSFERASE9 (GALT9) to the transverse cell walls. GALT9 is upregulated in the absence of miR775 – which is suppressed by stabilized HY5. This polarisation of pectin increases the elastic modulus (measured using Atomic Force Microscopy) of transverse cell walls in a wild-type system in response to light, therefore inhibiting elongation of the hypocotyl. These results offer new mechanical insight into the inhibition of hypocotyl growth in response to light, unveiling pectin and GALT9 as key players in this process. (Summary by Kes Maio @kesmaio.bsky.social) Curr. Biol. 10.1016/j.cub.2024.12.026
Light is a powerful signal, shaping plant development and growth. However, the cellular mechanisms that translate light signals into precise developmental responses are still being unravelled. The Arabidopsis hypocotyl (the embryonic stem, situated underneath the cotyledons in seedlings) rapidly restricts elongation upon light exposure. Zhang et al. combined molecular and mechanical techniques to uncover a succinct model for how this inhibition occurs in the dark-to-light transition. Initially, using time-lapse photography, they identified a key regulator inhibiting hypocotyl elongation: ELONGATED HYPOCOTYL 5 (HY5). Through chemical quantification and immunohistochemical analysis of the cell walls they found that, unlike in wild-type plants, there is no change in the accumulation of pectin in the hy5 mutant. Further, Raman microscopy showed that pectin is polarised to transverse walls of cells following light treatment – a pattern absent in hy5. This polarisation of pectin is due to the asymmetric localisation of GALACTOTRANSFERASE9 (GALT9) to the transverse cell walls. GALT9 is upregulated in the absence of miR775 – which is suppressed by stabilized HY5. This polarisation of pectin increases the elastic modulus (measured using Atomic Force Microscopy) of transverse cell walls in a wild-type system in response to light, therefore inhibiting elongation of the hypocotyl. These results offer new mechanical insight into the inhibition of hypocotyl growth in response to light, unveiling pectin and GALT9 as key players in this process. (Summary by Kes Maio @kesmaio.bsky.social) Curr. Biol. 10.1016/j.cub.2024.12.026
Brassinosteroids positively regulate growth through asymmetrical division
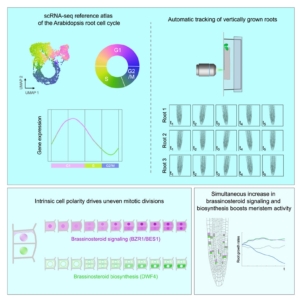 Brassinosteroids are plant hormones that play important roles in cell proliferation and expansion, but it is still unclear exactly what those roles are. Vukasinovic and colleagues show, in Arabidopsis thaliana root cells, that instrinsic brassinosteroid gradients signal anticlinal division. Single-cell RNA sequencing and vertical microscopy with automatic root tracking were used to observe brassinosteroid activity via both fluorescence and transcription in individual cells as the root progressed through the cell cycle. During the G1 phase, brassinosteroid activity increases, and the uneven distribution of the brassinosteroid signaling components leads to assymetric cell division, resulting in one brassinosteroid-active cell and one supporting cell. Using this information, a computational modeling simulation of growth in the root meristem was created to understand the importance of this asymmetrical division. By having the one daughter cell highly express brassinosteroids and the other express them on a delay, the process avoids negative feedback between signaling and biosynthesis and allows increased cell proliferation in the meristem. This work represents a major step forward in understanding brassinosteroid-regulated growth. Additionally, the root cell cycle reference and markers developed from the scRNA-seq data could inform many future experiments, even beyond brassinosteroids. (Summary by Elise Krespan) Cell 10.1016/j.cell.2025.02.011
Brassinosteroids are plant hormones that play important roles in cell proliferation and expansion, but it is still unclear exactly what those roles are. Vukasinovic and colleagues show, in Arabidopsis thaliana root cells, that instrinsic brassinosteroid gradients signal anticlinal division. Single-cell RNA sequencing and vertical microscopy with automatic root tracking were used to observe brassinosteroid activity via both fluorescence and transcription in individual cells as the root progressed through the cell cycle. During the G1 phase, brassinosteroid activity increases, and the uneven distribution of the brassinosteroid signaling components leads to assymetric cell division, resulting in one brassinosteroid-active cell and one supporting cell. Using this information, a computational modeling simulation of growth in the root meristem was created to understand the importance of this asymmetrical division. By having the one daughter cell highly express brassinosteroids and the other express them on a delay, the process avoids negative feedback between signaling and biosynthesis and allows increased cell proliferation in the meristem. This work represents a major step forward in understanding brassinosteroid-regulated growth. Additionally, the root cell cycle reference and markers developed from the scRNA-seq data could inform many future experiments, even beyond brassinosteroids. (Summary by Elise Krespan) Cell 10.1016/j.cell.2025.02.011
SHUKR in the sporophytic tissue directs male gametophyte development in Arabidopsis
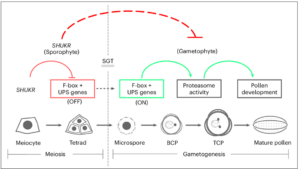 In flowering plants, reproduction involves the alternation of two generations: the diploid sporophyte and the haploid gametophyte. Traditionally, the male gametophyte was thought to develop autonomously with minimal regulation by the sporophyte. However, recent research by Sivakumar, Pandey, and Ramesha et al. challenges this view, identifying the SHUKR (SKR) gene in Arabidopsis, which plays a crucial role in male gametophyte development through the ubiquitin-proteasome system. The Arabidopsis SHUKR gene encodes a 119 amino acid protein present only in eudicots, but with no homology to other protein families. Using proSKR:GFP-gSKR reporter lines, SKR protein was detected in male meiocytes from prophase to the tetrad stage, suggesting its function in halting the gametophytic developmental program. The authors performed RNA-sequencing of meiotic anthers to reveal that misregulation of F-box genes leads to premature expression of gametogenesis-related proteins, causing defective pollen development and reduced fertility in the skr-1 mutants. This study also highlights the rapid evolution of the SHUKR protein family in eudicots, suggesting its adaptive role in gametophyte development across diverse plant species. The intense selective pressure on SHUKR and its regulatory control over F-box genes underscore the importance of protein turnover in male gametophyte diversification. These findings challenge the long-held paradigm of male gametophyte autonomy, providing compelling evidence that sporophytic regulation, influenced by internal and environmental factors, actively shapes gametophyte development. This research offers novel insights into plant reproductive strategies and evolutionary biology. (Summary by Gourav Arora @gourav_arora_g) Nature Plants 10.1038/s41477-025-01932-y
In flowering plants, reproduction involves the alternation of two generations: the diploid sporophyte and the haploid gametophyte. Traditionally, the male gametophyte was thought to develop autonomously with minimal regulation by the sporophyte. However, recent research by Sivakumar, Pandey, and Ramesha et al. challenges this view, identifying the SHUKR (SKR) gene in Arabidopsis, which plays a crucial role in male gametophyte development through the ubiquitin-proteasome system. The Arabidopsis SHUKR gene encodes a 119 amino acid protein present only in eudicots, but with no homology to other protein families. Using proSKR:GFP-gSKR reporter lines, SKR protein was detected in male meiocytes from prophase to the tetrad stage, suggesting its function in halting the gametophytic developmental program. The authors performed RNA-sequencing of meiotic anthers to reveal that misregulation of F-box genes leads to premature expression of gametogenesis-related proteins, causing defective pollen development and reduced fertility in the skr-1 mutants. This study also highlights the rapid evolution of the SHUKR protein family in eudicots, suggesting its adaptive role in gametophyte development across diverse plant species. The intense selective pressure on SHUKR and its regulatory control over F-box genes underscore the importance of protein turnover in male gametophyte diversification. These findings challenge the long-held paradigm of male gametophyte autonomy, providing compelling evidence that sporophytic regulation, influenced by internal and environmental factors, actively shapes gametophyte development. This research offers novel insights into plant reproductive strategies and evolutionary biology. (Summary by Gourav Arora @gourav_arora_g) Nature Plants 10.1038/s41477-025-01932-y
Classic regulators, new functions: FT and TFL1 shape Arabidopsis seed traits
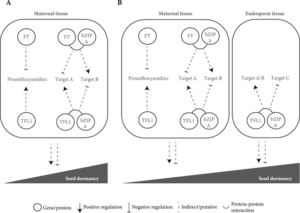 FLOWERING LOCUS T (FT) and TERMINAL FLOWER1 (TFL1), two members of the phosphatidylethanolamine-binding protein (PEBP) family, are well-known regulators of flowering time and inflorescence architecture in Arabidopsis thaliana. However, recent work by Bigas et al. uncovers their additional roles in regulating seed traits, such as dormancy and size. Using CRISPR/Cas9-generated mutants in the Ler background, the study shows that tfl1 mutants produce larger seeds with reduced dormancy, while ft mutants exhibit increased dormancy with minimal changes in seed size. Reporter assays reveal that both proteins co-localize in maternal vascular tissues, but only TFL1 is present in the endosperm, suggesting tissue-specific roles. TFL1, but not FT, influences seed size, potentially by delaying endosperm cellularization. The authors propose two hypotheses: one in which FT and TFL1 act together in a shared pathway affecting proanthocyanidin biosynthesis in maternal tissues, and another in which they function independently due to their distinct expression patterns. These findings expand the known functions of FT and TFL1, emphasizing the importance of revisiting classic developmental regulators in the context of non-canonical traits, with broad implications for crop improvement and agricultural productivity. (Summary by Gourav Arora @gourav_arora_g) J. Exp. Bot 10.1093/jxb/erae466
FLOWERING LOCUS T (FT) and TERMINAL FLOWER1 (TFL1), two members of the phosphatidylethanolamine-binding protein (PEBP) family, are well-known regulators of flowering time and inflorescence architecture in Arabidopsis thaliana. However, recent work by Bigas et al. uncovers their additional roles in regulating seed traits, such as dormancy and size. Using CRISPR/Cas9-generated mutants in the Ler background, the study shows that tfl1 mutants produce larger seeds with reduced dormancy, while ft mutants exhibit increased dormancy with minimal changes in seed size. Reporter assays reveal that both proteins co-localize in maternal vascular tissues, but only TFL1 is present in the endosperm, suggesting tissue-specific roles. TFL1, but not FT, influences seed size, potentially by delaying endosperm cellularization. The authors propose two hypotheses: one in which FT and TFL1 act together in a shared pathway affecting proanthocyanidin biosynthesis in maternal tissues, and another in which they function independently due to their distinct expression patterns. These findings expand the known functions of FT and TFL1, emphasizing the importance of revisiting classic developmental regulators in the context of non-canonical traits, with broad implications for crop improvement and agricultural productivity. (Summary by Gourav Arora @gourav_arora_g) J. Exp. Bot 10.1093/jxb/erae466