Plant Science Research Weekly: April 1, 2022
Chloroplast protein import determines plant proteostasis and retrograde signaling
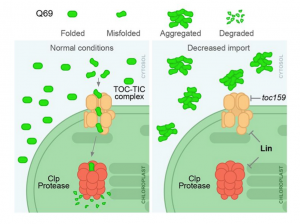 The neurological condition Huntington’s disease is caused by mutated versions of the Huntingtin gene that encode expanded stretches of polyglutamine repeats, resulting in protein aggregation. However, despite the widespread presence of polyglutamine repeat regions in plant proteomes, similar pathologies have yet to be observed. To investigate this paradox, Llamas and colleagues expressed aggregation-prone fragments of the human Huntingtin protein, Q28 and Q69, in Arabidopsis thaliana plants. Amazingly the protein did not aggregate and as a result the plants remained healthy. Further protein interaction studies revealed that Q28 and Q69 are imported into the chloroplast and degraded by the stromal processing peptidase (SPP) protein. The researchers then co-expressed a version of the Arabidopsis SPP protein in a human cell line also expressing a polyglutamine protein. Remarkably these co-expression cells showed reduced aggregation of the protein, suggesting a potential therapeutic avenue for the treatment of these diseases in humans. The group also showed that inhibition of chloroplast protein import not only led to the aggregation of Q69, but also of an endogenous plant polyglutamine containing protein, plastid casein kinase 2. Under these conditions, the protein forms cytosolic aggregates and accumulates in the nucleus, suggesting a potential role for polyglutamate-containing proteins in plant organellar retrograde signaling. (Summary by Rory Burke @rorby95) bioRxiv 10.1101/2022.03.19.484971 (And don’t miss the animated Tweetorial created by the first author).
The neurological condition Huntington’s disease is caused by mutated versions of the Huntingtin gene that encode expanded stretches of polyglutamine repeats, resulting in protein aggregation. However, despite the widespread presence of polyglutamine repeat regions in plant proteomes, similar pathologies have yet to be observed. To investigate this paradox, Llamas and colleagues expressed aggregation-prone fragments of the human Huntingtin protein, Q28 and Q69, in Arabidopsis thaliana plants. Amazingly the protein did not aggregate and as a result the plants remained healthy. Further protein interaction studies revealed that Q28 and Q69 are imported into the chloroplast and degraded by the stromal processing peptidase (SPP) protein. The researchers then co-expressed a version of the Arabidopsis SPP protein in a human cell line also expressing a polyglutamine protein. Remarkably these co-expression cells showed reduced aggregation of the protein, suggesting a potential therapeutic avenue for the treatment of these diseases in humans. The group also showed that inhibition of chloroplast protein import not only led to the aggregation of Q69, but also of an endogenous plant polyglutamine containing protein, plastid casein kinase 2. Under these conditions, the protein forms cytosolic aggregates and accumulates in the nucleus, suggesting a potential role for polyglutamate-containing proteins in plant organellar retrograde signaling. (Summary by Rory Burke @rorby95) bioRxiv 10.1101/2022.03.19.484971 (And don’t miss the animated Tweetorial created by the first author).
A synthetic switch based on orange carotenoid protein to control blue-green light responses in chloroplasts
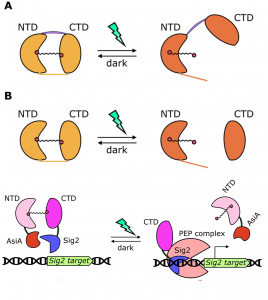 Synthetic biology aims to engineer and redesign components of natural organisms for useful purposes. One of the most prolific areas of synthetic biology is based on the engineering of photoreactive proteins with signaling potential, such as photoreceptors. Natural photoreceptors consist of a prosthetic chromophore which is responsible for light perception, and an apoprotein, which transduces the light signal. To expand beyond the natural existing photoreceptors, researchers have been developing synthetic ones by interchanging interesting protein domains. In this work, Piccinini et al. developed a light-dependent, chloroplast localized photoswitch system based on cyanobacterial Orange Carotenoid Proteins (OCPs), which use a keto-carotenoid as a chromophore. OCPs have distinct N- and C- terminal domains (NTD and CTD) that change conformation depending on light perception; light induces the domains to move away from each other. As a proof of concept, the authors engineered a synthetic photoswitch composed of two polypeptides, each encoding the OCP NTD or CDT fused to half of a NanoLuc luciferase protein to serve as a reporter. In darkness, the OCP domains interact and light is emitted from NanoLuc, but upon illumination the domains move apart and no light is emitted. (Because Arabidopsis does not produce the keto-carotenoid required for OCP activation, it is also necessary to introduce a keto-carotenoid biosynthetic enzyme into plants for the photoswitch to function).The authors next showed that this system could be used for light-regulated transcriptional control, by fusing the OCP NTD to the N-terminus of the T4 phage anti-sigma factor AsiA and the CTD to the C-terminus of Arabidopsis Sigma2 (Sig2). In the dark, Sig2 target is not transcribed, but when blue-green light is applied, the Sig2 target ATPI is transcribed. This work enlarges the toolbox for synthetic biologists by means of OCPs to fine tune the expression levels of target genes upon exposure to specific light cues. (Summary by Eva Maria Gomez Alvarez, @eva_ga96). Plant Physiol. 10.1093/plphys/kiac122
Synthetic biology aims to engineer and redesign components of natural organisms for useful purposes. One of the most prolific areas of synthetic biology is based on the engineering of photoreactive proteins with signaling potential, such as photoreceptors. Natural photoreceptors consist of a prosthetic chromophore which is responsible for light perception, and an apoprotein, which transduces the light signal. To expand beyond the natural existing photoreceptors, researchers have been developing synthetic ones by interchanging interesting protein domains. In this work, Piccinini et al. developed a light-dependent, chloroplast localized photoswitch system based on cyanobacterial Orange Carotenoid Proteins (OCPs), which use a keto-carotenoid as a chromophore. OCPs have distinct N- and C- terminal domains (NTD and CTD) that change conformation depending on light perception; light induces the domains to move away from each other. As a proof of concept, the authors engineered a synthetic photoswitch composed of two polypeptides, each encoding the OCP NTD or CDT fused to half of a NanoLuc luciferase protein to serve as a reporter. In darkness, the OCP domains interact and light is emitted from NanoLuc, but upon illumination the domains move apart and no light is emitted. (Because Arabidopsis does not produce the keto-carotenoid required for OCP activation, it is also necessary to introduce a keto-carotenoid biosynthetic enzyme into plants for the photoswitch to function).The authors next showed that this system could be used for light-regulated transcriptional control, by fusing the OCP NTD to the N-terminus of the T4 phage anti-sigma factor AsiA and the CTD to the C-terminus of Arabidopsis Sigma2 (Sig2). In the dark, Sig2 target is not transcribed, but when blue-green light is applied, the Sig2 target ATPI is transcribed. This work enlarges the toolbox for synthetic biologists by means of OCPs to fine tune the expression levels of target genes upon exposure to specific light cues. (Summary by Eva Maria Gomez Alvarez, @eva_ga96). Plant Physiol. 10.1093/plphys/kiac122
A conserved superlocus regulates above- and belowground root initiation
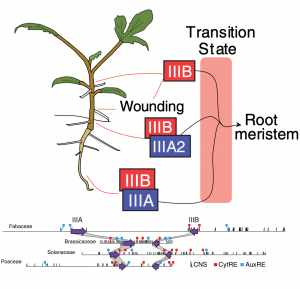 The vascular plant body composed of root and shoot is specified during embryogenesis, but most flowering plants can also develop additional root systems post-embryonically: lateral roots, as a response to wounding, or shoot-borne roots (“adventitious” roots, literally meaning “in the wrong place”). The initiation mechanism of these roots as well as their parent tissue and the ontogenic relationships between them are poorly understood. Since Arabidopsis thaliana rarely produces shoot-borne roots, Omary and colleagues used tomato (Solanum lycopersicum) to uncover the molecular basis of shoot-borne root initiation. Hormonal fluorescent reporters, single-cell transcriptomics and CRISPR null-mutants revealed that shoot-borne roots originate from differentiated phloem-associated cells that go through an uncharacterized transitory cell identity: from differentiated phloem to root meristem. Subsequent analysis pointed towards a LBD transcription factor whose null-mutant phenotype resulted in stems and hypocotyls completely lacking roots, thus was named SHOOTBORNE-ROOTLESS (SBRL). Strikingly, phylogenetic analysis of SBRL orthologs and loss-of-function mutants of potato (Solanum tuberosum) SBRL revealed that the regulation of the shoot-borne root initiation program by IIIB LBDs (a subclass of LBDs) is deeply conserved within angiosperms. In analyzing the genomic regions of IIIB LBDs orthologs, the authors found that they are almost always found in a “superlocus” that also contains IIIA LBDs and conserved auxin and cytokinin response cis elements. The double mutant sbrl bsbrl (BROTHER OF SHOOTBORNE-ROOTLESS, the syntenic IIIA LBD adjacent to SBRL) completely lacks shoot-borne roots and displays a significant reduction in lateral roots, indicating that IIIB and IIIA LBDs function in root initiation in different contexts. This fascinating paper describing how a deeply conserved angiosperm superlocus employs individual paralogs to regulate the initiation of postembryonic roots in different developmental contexts and is a valuable contribution for understanding the genetic programs underlying different types of root systems. (Summary by Jesus Leon @jesussaur) Science 10.1126/science.abf4368
The vascular plant body composed of root and shoot is specified during embryogenesis, but most flowering plants can also develop additional root systems post-embryonically: lateral roots, as a response to wounding, or shoot-borne roots (“adventitious” roots, literally meaning “in the wrong place”). The initiation mechanism of these roots as well as their parent tissue and the ontogenic relationships between them are poorly understood. Since Arabidopsis thaliana rarely produces shoot-borne roots, Omary and colleagues used tomato (Solanum lycopersicum) to uncover the molecular basis of shoot-borne root initiation. Hormonal fluorescent reporters, single-cell transcriptomics and CRISPR null-mutants revealed that shoot-borne roots originate from differentiated phloem-associated cells that go through an uncharacterized transitory cell identity: from differentiated phloem to root meristem. Subsequent analysis pointed towards a LBD transcription factor whose null-mutant phenotype resulted in stems and hypocotyls completely lacking roots, thus was named SHOOTBORNE-ROOTLESS (SBRL). Strikingly, phylogenetic analysis of SBRL orthologs and loss-of-function mutants of potato (Solanum tuberosum) SBRL revealed that the regulation of the shoot-borne root initiation program by IIIB LBDs (a subclass of LBDs) is deeply conserved within angiosperms. In analyzing the genomic regions of IIIB LBDs orthologs, the authors found that they are almost always found in a “superlocus” that also contains IIIA LBDs and conserved auxin and cytokinin response cis elements. The double mutant sbrl bsbrl (BROTHER OF SHOOTBORNE-ROOTLESS, the syntenic IIIA LBD adjacent to SBRL) completely lacks shoot-borne roots and displays a significant reduction in lateral roots, indicating that IIIB and IIIA LBDs function in root initiation in different contexts. This fascinating paper describing how a deeply conserved angiosperm superlocus employs individual paralogs to regulate the initiation of postembryonic roots in different developmental contexts and is a valuable contribution for understanding the genetic programs underlying different types of root systems. (Summary by Jesus Leon @jesussaur) Science 10.1126/science.abf4368
The urban environment led to unintended adaptive evolution in plants
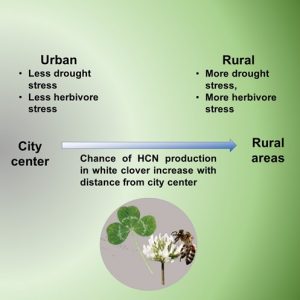 Generally, evolution is driven by natural selection, but not always. Human activities lead to the creation of unique niches, and other organisms must adapt accordingly. Cities are unique niches that are significantly different from rural areas and natural conditions. The urban habitat provides plants with adequate water and protection from herbivores. In a recent study, Santangelo et al. phenotyped over 110000 white clover plants from 6169 populations globally. These include many urban and nearby rural white clover populations. The authors sequenced 2074 of these populations. The authors observed a higher probability of hydrogen cyanide (HCN) production in white clover populations in a rural environment. The production of HCN is a stress response that is elicited by herbivores or drought conditions. In many cities, the tendency to produce HCN increases with distance from the city center. This indicates adaptive evolution driven by urban conditions. It will be interesting to find if there is gain of HCN production in rural areas or plants that are adapted to urban conditions gave up this feature. If you ask me, my bet is one the second one. (Summary by Kamal Kumar Malukani, @KamalMalukani) Science, 10.1126/science.abk0989
Generally, evolution is driven by natural selection, but not always. Human activities lead to the creation of unique niches, and other organisms must adapt accordingly. Cities are unique niches that are significantly different from rural areas and natural conditions. The urban habitat provides plants with adequate water and protection from herbivores. In a recent study, Santangelo et al. phenotyped over 110000 white clover plants from 6169 populations globally. These include many urban and nearby rural white clover populations. The authors sequenced 2074 of these populations. The authors observed a higher probability of hydrogen cyanide (HCN) production in white clover populations in a rural environment. The production of HCN is a stress response that is elicited by herbivores or drought conditions. In many cities, the tendency to produce HCN increases with distance from the city center. This indicates adaptive evolution driven by urban conditions. It will be interesting to find if there is gain of HCN production in rural areas or plants that are adapted to urban conditions gave up this feature. If you ask me, my bet is one the second one. (Summary by Kamal Kumar Malukani, @KamalMalukani) Science, 10.1126/science.abk0989
Climate shapes the seed germination niche of temperate flowering plants: a meta-analysis of European seed conservation data
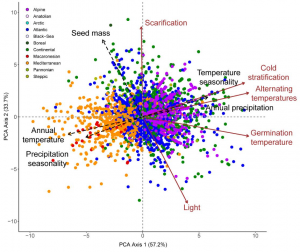 The seed germination niche is the set of environmental conditions in which a seed can germinate. This collection of requirements is expected to be tuned to the climate each species encounters in its natural habitat, but this hypothesis remains to be formally tested. Here, Carta and colleagues make use of the database from the European Native Seed Conservation Network (ENSCONET) to conduct the first meta-analysis of the seed germination niche on the continental scale. The study was focused on two main aspects: the correlation between species germination requirements and climate and the role of evolutionary history in this association. Overall, germination temperatures and responses to alternate temperature regimes and cold stratification were shaped by climate so that species from warm and seasonally dry environments have higher germinability when exposed to cool, constant temperatures but no cold stratification. However, some relationships between germination requirements and climate were only found in certain clades. For example, warm stratification promoted the germination of commelinids from humid climates. This fascinating study nicely illustrates the prominent role of climate in constructing the seed germination niche and the importance of evolutionary history in this process. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Ann. Bot. 10.1093/aob/mcac037
The seed germination niche is the set of environmental conditions in which a seed can germinate. This collection of requirements is expected to be tuned to the climate each species encounters in its natural habitat, but this hypothesis remains to be formally tested. Here, Carta and colleagues make use of the database from the European Native Seed Conservation Network (ENSCONET) to conduct the first meta-analysis of the seed germination niche on the continental scale. The study was focused on two main aspects: the correlation between species germination requirements and climate and the role of evolutionary history in this association. Overall, germination temperatures and responses to alternate temperature regimes and cold stratification were shaped by climate so that species from warm and seasonally dry environments have higher germinability when exposed to cool, constant temperatures but no cold stratification. However, some relationships between germination requirements and climate were only found in certain clades. For example, warm stratification promoted the germination of commelinids from humid climates. This fascinating study nicely illustrates the prominent role of climate in constructing the seed germination niche and the importance of evolutionary history in this process. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Ann. Bot. 10.1093/aob/mcac037
Seed dormancy in space and time: global distribution, paleo- and present climatic drivers and evolutionary adaptations ($)
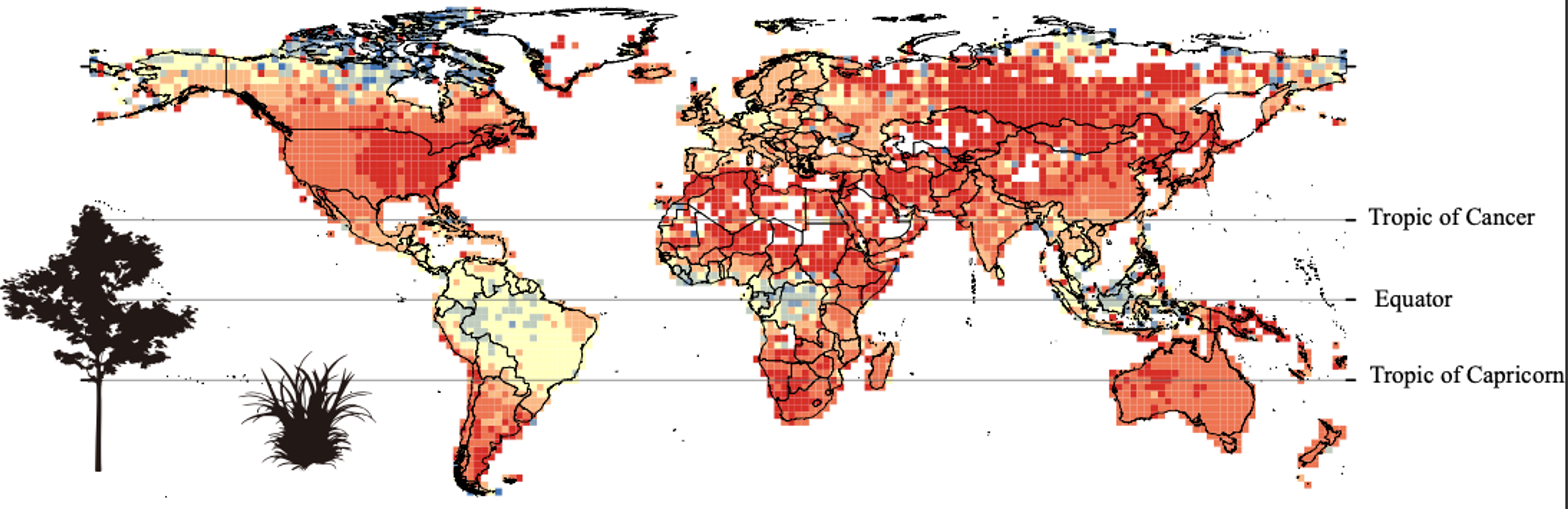 Seed dormancy is widely recognized as a key mechanism to ensure that germination takes place under the most suitable conditions. Such is its importance that multiple studies have described the morphological, physiological, and genetic mechanisms behind it, yet its global distribution and the past and current environmental drivers behind it have seldom been addressed. Based on a database of more than 12,400 species, Zhang, Liu, Sun, and colleagues found that the lowest proportion of species with dormant seeds is found in the Arctic and the tropical rainforests near the Equator, whereas the middle-latitudes host the highest proportion, and areas with seasonal climates and wide diurnal temperature ranges have a high proportion of species with dormant seeds. The authors also found that this proportion has gradually decreased since the Cenozoic –the era between 65 million years ago and the present– potentially due to increasing temperatures that favor the absence of dormancy. However, all these global patterns varied between herbaceous and woody species, implying that the adaptive value of seed dormancy depends on plant growth form. This research provides fascinating insights into the biogeography and evolution of seed dormancy. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) New Phytol. 10.1111/nph.18099
Seed dormancy is widely recognized as a key mechanism to ensure that germination takes place under the most suitable conditions. Such is its importance that multiple studies have described the morphological, physiological, and genetic mechanisms behind it, yet its global distribution and the past and current environmental drivers behind it have seldom been addressed. Based on a database of more than 12,400 species, Zhang, Liu, Sun, and colleagues found that the lowest proportion of species with dormant seeds is found in the Arctic and the tropical rainforests near the Equator, whereas the middle-latitudes host the highest proportion, and areas with seasonal climates and wide diurnal temperature ranges have a high proportion of species with dormant seeds. The authors also found that this proportion has gradually decreased since the Cenozoic –the era between 65 million years ago and the present– potentially due to increasing temperatures that favor the absence of dormancy. However, all these global patterns varied between herbaceous and woody species, implying that the adaptive value of seed dormancy depends on plant growth form. This research provides fascinating insights into the biogeography and evolution of seed dormancy. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) New Phytol. 10.1111/nph.18099
An oomycete peptide cytolysin forms transient small pores in lipid membranes
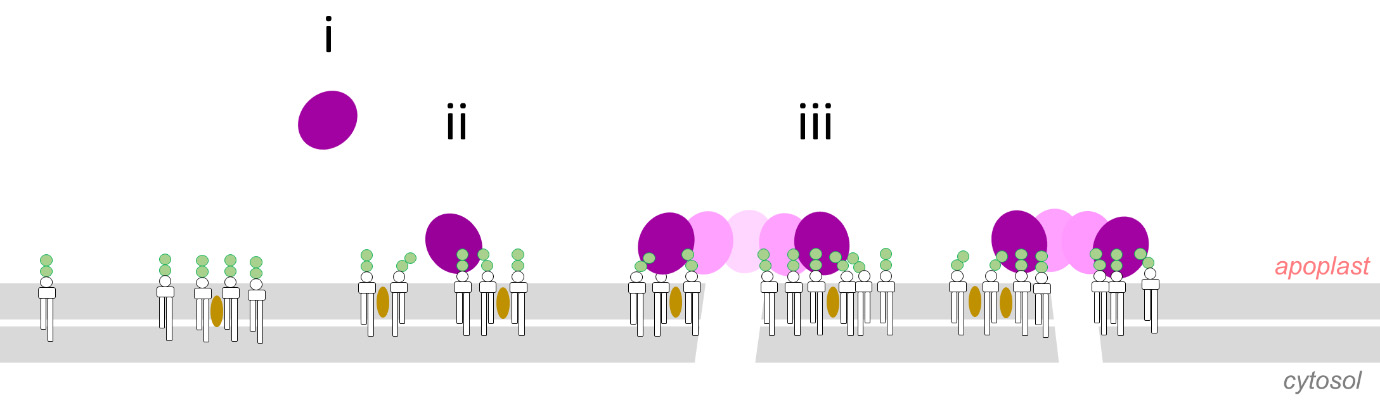 NLPs (Necrosis and ethylene-inducing peptide 1–like proteins) are small peptides produced by a variety of plant pathogens. Some NLPs are cytolysins meaning that they trigger lysis of their target cells. Here, Pirc et al. use an assortment of tools including molecular dynamics (MD) simulations, neutron reflectometry (NR), and giant unilamellar vesicles (GUVs) to identify how one of these, NLPPya from the phytopathogenic oomycete Pythium aphanidermatum, interacts with and damages plant cell membranes. Previous studies found that NLPs (shown in purple in the image) preferentially interact with the polar headgroup of glycosylinositol phosphorylceramides (GIPCs; shown in green in the image) at the surface of the plasma membrane through electrostatic interactions, and this interaction is strengthened by the presence of sterols (tan in the image). NLPPya interacts with multiple GIPCs, leading to the formation of aggregates and the opening of small, transient pores in the lipid bilayer. Interestingly this mode of action, in which the NLP sits at the surface of the membane, is different from that of other pore-forming toxins such as those produced by Bacillus thuringiensis (Bt toxin)or Bacillus anthracis (anthrax) which directly insert into the lipid bilayer to produce a pore. These findings will be helpful in developing strategies to combat NLP-producing pathogens. (Summary by Mary Williams @PlantTeaching) Sci. Adv. 10.1126/sciadv.abj9406
NLPs (Necrosis and ethylene-inducing peptide 1–like proteins) are small peptides produced by a variety of plant pathogens. Some NLPs are cytolysins meaning that they trigger lysis of their target cells. Here, Pirc et al. use an assortment of tools including molecular dynamics (MD) simulations, neutron reflectometry (NR), and giant unilamellar vesicles (GUVs) to identify how one of these, NLPPya from the phytopathogenic oomycete Pythium aphanidermatum, interacts with and damages plant cell membranes. Previous studies found that NLPs (shown in purple in the image) preferentially interact with the polar headgroup of glycosylinositol phosphorylceramides (GIPCs; shown in green in the image) at the surface of the plasma membrane through electrostatic interactions, and this interaction is strengthened by the presence of sterols (tan in the image). NLPPya interacts with multiple GIPCs, leading to the formation of aggregates and the opening of small, transient pores in the lipid bilayer. Interestingly this mode of action, in which the NLP sits at the surface of the membane, is different from that of other pore-forming toxins such as those produced by Bacillus thuringiensis (Bt toxin)or Bacillus anthracis (anthrax) which directly insert into the lipid bilayer to produce a pore. These findings will be helpful in developing strategies to combat NLP-producing pathogens. (Summary by Mary Williams @PlantTeaching) Sci. Adv. 10.1126/sciadv.abj9406
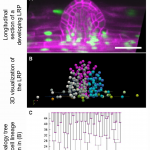
Live Plant Cell Tracking: A Fiji plugin to analyze cell proliferation dynamics and understand morphogenesis
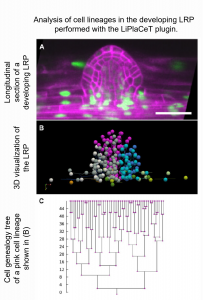 Morphogenesis in plants occurs in 3D space over time, referred to as 4D. Recent advances in high-resolution microscopy imaging and computational pipelines have helped to better understand plant organ development at cellular resolution. Hernández-Herrera et al. developed a plugin, “Live Plant Cell Tracking” (LiPlaCeT),” which allows the analysis of 4D data to extract quantitative cellular parameters underlying organogenesis. The Arabidopsis thaliana primary root apical meristem (RAM) and lateral root primordia (LRPs) were analyzed with LiPlaCeT. The authors focused on the analysis of cell cycle duration in the LRP central and longitudinally flanking domains. Their findings showed that the cell cycle number in the flanking domain was two-fold lower than in the central one. In addition, they performed analysis in the whole RAM region, extracting data such as distance to the initial nucleus, the distance between nuclei, cell growth, cell displacement, and proliferation activity. The authors show that LiPlaCeT is another way to analyze the 4D complexity of plant organs in detail, which will help the scientific community to understand critical developmental processes during plant morphogenesis. (Summary by Andrea Gómez Felipe @andreagomezfe) Plant Physiol. 10.1093/plphys/kiab530
Morphogenesis in plants occurs in 3D space over time, referred to as 4D. Recent advances in high-resolution microscopy imaging and computational pipelines have helped to better understand plant organ development at cellular resolution. Hernández-Herrera et al. developed a plugin, “Live Plant Cell Tracking” (LiPlaCeT),” which allows the analysis of 4D data to extract quantitative cellular parameters underlying organogenesis. The Arabidopsis thaliana primary root apical meristem (RAM) and lateral root primordia (LRPs) were analyzed with LiPlaCeT. The authors focused on the analysis of cell cycle duration in the LRP central and longitudinally flanking domains. Their findings showed that the cell cycle number in the flanking domain was two-fold lower than in the central one. In addition, they performed analysis in the whole RAM region, extracting data such as distance to the initial nucleus, the distance between nuclei, cell growth, cell displacement, and proliferation activity. The authors show that LiPlaCeT is another way to analyze the 4D complexity of plant organs in detail, which will help the scientific community to understand critical developmental processes during plant morphogenesis. (Summary by Andrea Gómez Felipe @andreagomezfe) Plant Physiol. 10.1093/plphys/kiab530