Petunia Battlefield in Style: S-RNases vs. SLF proteins
Sun et al. use CRISPR/Cas9 in Petunia to establish the essential role of SLF proteins in self-compatibility and reveal their complex interactions with S-RNases. Plant Cell https://doi.org/10.1105/tpc.18.00615.
By Linhan Sun and Teh-hui Kao, The Pennsylvania State University
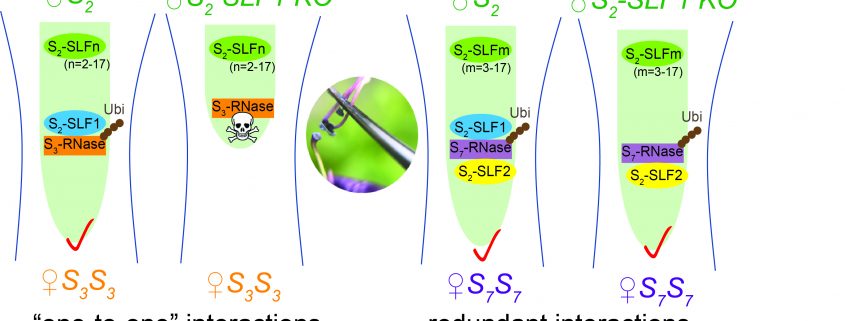
Background: Plant inbreeding results in reduced fitness of the progeny. Since plants cannot move around to select mates, many flowering plants have adopted a reproductive strategy, self-incompatibility, that allows pistils to prevent inbreeding by rejecting genetically identical self pollen and only accepting non-self pollen for fertilization. The Petunia pistil uses S-RNase as a toxin to reject self pollen. Non-self pollen is thought to escape S-RNase toxicity using a suite of its own proteins, S-locus F-box (SLF) proteins, to meditate non-self S-RNase degradation. For example, pollen having the S2-haplotype contains 17 types of SLF protein. Each type can mediate the degradation of some non-self S-RNases from pistils with S-haplotypes other than S2. Collectively, the entire suite of SLF proteins can detoxify all non-self S-RNases.
Question: We wished to determine whether pollen indeed requires SLF proteins to detoxify non-self S-RNases when a pollen tube grows in a genetically compatible pistil. If an SLF is essential for detoxifying a certain S-RNase, pollen lacking this SLF should be rejected by the pistil producing this S-RNase.
Findings: We used CRISPR/Cas9 genome-editing to generate insertion/deletion mutations in the S2-SLF1 gene of S2-haplotype of Petunia inflata to create mutant plants whose pollen did not produce a functional S2-SLF1. Mutant S2 pollen was rejected by pistils producing S3-RNase or S13-RNase but was still accepted by pistils producing S7-RNase or S12-RNase. We separately expressed each of the 17 SLF proteins of S2-haplotype in pollen of various S-haplotypes and showed that only S2-SLF1 could detoxify S3-RNase, whereas in addition to S2-SLF1, S2-SLF2 and S2-SLF5 could also detoxify S7-RNase and S12-RNase, respectively. Therefore, S2-SLF1 is essential in the defense of S2 pollen against S3-RNase, and S2 pollen uses multiple SLF proteins to defend against some other S-RNases as a fail-safe strategy.
Next steps: The next challenge is to understand why among the 17 SLF proteins produced by S2 pollen, only one, or a few, can detoxify a particular S-RNase. At the biochemical level, which amino acids are responsible for enabling an SLF to interact with certain S-RNase(s), and at the structural level, how does it interact with them?
Linhan Sun, Justin S. Williams, Shu Li, Lihua Wu, Wasi A. Khatri, Patrick G. Stone, Matthew D. Keebaugh, Teh-hui Kao. (2018). S-Locus F-Box Proteins Are Solely Responsible for S-RNase-Based Self-Incompatibility of Petunia Pollen. Plant Cell 30: 2959-2972; DOI: https://doi.org/10.1105/tpc.18.00615
Key words: CRISPR/Cas9, Petunia, Pollen-Pistil Interactions, Self-Incompatibility