PeTriBERT : Augmenting BERT with tridimensional encoding for inverse protein folding and design (bioRxiv)
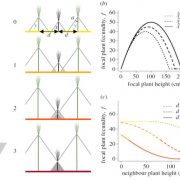
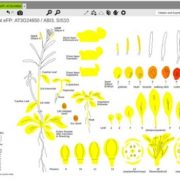
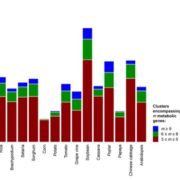
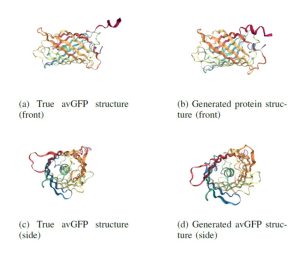 The AlphaFoldDB database recently made headlines by predicting the three-dimensional protein structure of millions of proteins based on their primary amino acid sequence. Most of these models remain to be tested, but it’s a place to start. Here, Dumortier et al. add an important complementary tool for improving our ability to correlate protein primary sequence and tertiary structure, in a tool that goes the other way; inverse protein folding. They trained their model on more than 350,000 protein sequences retrieved from the AlphaFoldDB. This model is an extension of BERT (Bidirectional Encoder Representations from Transformers), which is a machine learning / AI tool developed for language and grammar tasks (think about how Word identifies spelling and grammar mistakes in your writing). PeTriBERT (PeTriBERT: Proteins embedded in Tridimensional representation in a BERT model) enabled the researchers to generate sequences that fold to resemble GFP, of which 9/10 show no amino acid sequence homology to GFP. This tool then will enable researchers to design proteins with specific structures (and possibly similar catalytic or functional properties) but totally unique primary sequences; in silico evolution of synthetic proteins. Like so much of AI, the potential applications of this inverse-folding approach are stunning. (Summary by Mary Williams @PlantTeaching) bioRxiv 10.1101/2022.08.10.503344
The AlphaFoldDB database recently made headlines by predicting the three-dimensional protein structure of millions of proteins based on their primary amino acid sequence. Most of these models remain to be tested, but it’s a place to start. Here, Dumortier et al. add an important complementary tool for improving our ability to correlate protein primary sequence and tertiary structure, in a tool that goes the other way; inverse protein folding. They trained their model on more than 350,000 protein sequences retrieved from the AlphaFoldDB. This model is an extension of BERT (Bidirectional Encoder Representations from Transformers), which is a machine learning / AI tool developed for language and grammar tasks (think about how Word identifies spelling and grammar mistakes in your writing). PeTriBERT (PeTriBERT: Proteins embedded in Tridimensional representation in a BERT model) enabled the researchers to generate sequences that fold to resemble GFP, of which 9/10 show no amino acid sequence homology to GFP. This tool then will enable researchers to design proteins with specific structures (and possibly similar catalytic or functional properties) but totally unique primary sequences; in silico evolution of synthetic proteins. Like so much of AI, the potential applications of this inverse-folding approach are stunning. (Summary by Mary Williams @PlantTeaching) bioRxiv 10.1101/2022.08.10.503344