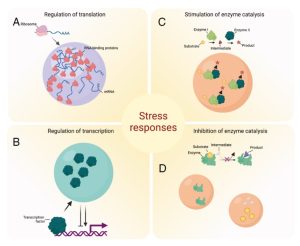
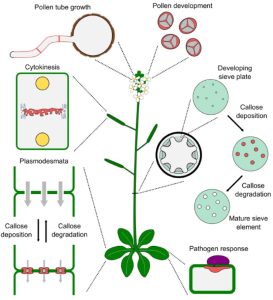
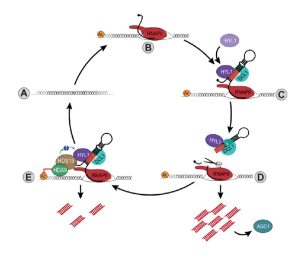
Current Position:
Scientist, Syngenta Crop Protection, LLC, Research Triangle Park, NC 27709
Education:
Ph.D., Horticulture, Purdue University, West Lafayette, Indiana, USA
M.S., Plant Biology, Southern Illinois University, Carbondale, Illinois, USA
B.S., Seed Science Technology, Osmania University, Hyderabad, India
Non-scientific Interests:
Reading and watching science fiction, travelling, cooking, and spending time with my family
Brief bio:
My path to become a Plant Biologist was serendipitous. Initially planning to earn a degree in Biology and specializing in microbiology, I enrolled into a B.S. Biology program at a college affiliated to Osmania University, Hyderabad, India. Unfortunately, that year the Biology Department experienced a spate of retirements, which resulted in a staffing crisis for teaching foundational courses. Not satisfied with the teaching quality and in fear of having to start a new program elsewhere, I made a switch mid-semester to an agriculture-based B.S. program. After graduating at the top of my class, I came to the US for an M.S program in Plant Biology at SIU Carbondale, Illinois. I owe my initial training as a Plant Physiologist to (the late) Dr. Stephen Ebbs, in his lab I investigated arsenic transport and speciation in rice, Indian mustard, and hyperaccumulating ferns. I expanded my skills as a molecular biologist during my Ph.D. program in Dr. K.G Raghothama’s lab at Purdue University. There, I characterized transcriptional programs key to plant phosphate signaling responses that led to several well-cited publications.
My interest in “omics” approaches (especially genomics and transcriptomics) and their impact on plant biology brought me to Dr. Pam Green’s lab at the University of Delaware for my postdoctoral work. I am thankful to Dr. Green for my training in NGS approaches and deeper understanding of plant RNA biology. Working with the RNA degradome that provides a genome-wide snapshot of RNA decay intermediates, introduced me to bioinformatics and analysis of large datasets. Using this approach, I helped identify substrates and novel associations of cytoplasmic exoribonuclease AtXRN4, and more recently mRNAs targeted by endoribonuclease AtDNE1.
As an Associate Scientist in the Green lab, I had a major role in the development, writing and execution of the National Science Foundation grant that funded our work identifying substrates of Arabidopsis endoribonucleases. I also took up the opportunity to train and mentor young scientists. Two especially talented students, Catherine Stuart (co-author, M.S) and undergraduate Anna DiBattista (currently Ph.D. student at UNC Chapel Hill) were instrumental in the characterization of AtDNE1.
Currently, I am using my dual expertise in plant molecular biology and bioinformatics to characterize transgenic events in crop plants at Syngenta.

 Wenjiao Zou, first author of “Lipid transport protein ORP2A promotes glucose signaling by facilitating RGS1 degradation”
Wenjiao Zou, first author of “Lipid transport protein ORP2A promotes glucose signaling by facilitating RGS1 degradation”

 Yuxin Shen, first author of
Yuxin Shen, first author of 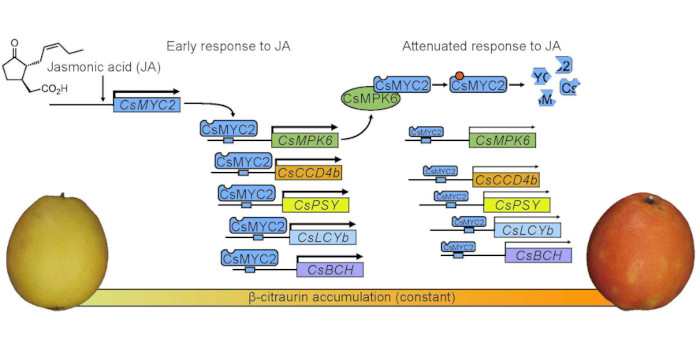 Findings: We found that exogenous methyl jasmonate (MeJA) treatment of ‘Newhall’ orange fruit promoted β-citraurin accumulation and fruit peel coloration. The jasmonate signaling master transcription factor CsMYC2 bound promoters of β-citraurin biosynthetic genes and activated their expression and promoted expression of the MAP kinase gene CsMPK6. In turn, CsMPK6 interacted with CsMYC2 to decrease the promoter binding activity of CsMYC2 to its target promoters. CsMPK6 also phosphorylated CsMYC2 to accelerate its degradation and thus, attenuated the jasmonate response for citrus to prevent the fruit from overreacting to jasmonates.
Findings: We found that exogenous methyl jasmonate (MeJA) treatment of ‘Newhall’ orange fruit promoted β-citraurin accumulation and fruit peel coloration. The jasmonate signaling master transcription factor CsMYC2 bound promoters of β-citraurin biosynthetic genes and activated their expression and promoted expression of the MAP kinase gene CsMPK6. In turn, CsMPK6 interacted with CsMYC2 to decrease the promoter binding activity of CsMYC2 to its target promoters. CsMPK6 also phosphorylated CsMYC2 to accelerate its degradation and thus, attenuated the jasmonate response for citrus to prevent the fruit from overreacting to jasmonates.
 Catherine Stuart, co-first author of
Catherine Stuart, co-first author of  Vinay Nagarajan, co-first author of
Vinay Nagarajan, co-first author of