Recently, we’ve been profiling first authors of Plant Cell papers that are selected for In Brief summaries. Here are the first-author profiles from the December issue of The Plant Cell.
 Current Position: Postdoctoral fellow in Agricultural Biotechnology Research Center, Academia Sinica, Taiwan.
Current Position: Postdoctoral fellow in Agricultural Biotechnology Research Center, Academia Sinica, Taiwan.
Education: PhD: TIGP-MBAS, Graduate Institute of Biotechnology, National Chung Hsing University (NCHU), Taiwan.
Non-scientific Interests: Hiking and watching movies.
After obtaining a B.S. degree in Botany at NCHU, I started my journey in plant research by joining Dr. Bor-yaw Lin’s lab as an M.S. student at the Institute of Molecular Biology of the same school. In his lab, I learned cytogenetics by working on the propagation of maize multiple-mutants, observation of B chromosomes, and molecular biological assays. After graduation, I spent a couple of years serving as a teaching assistant in a Genetics class at the same institute and helped to handle administrative affairs. To continue to pursue my interest in plant research, I joined the Ph.D. program in Molecular and Biological Agricultural Sciences (Taiwan International Graduate Program) co-operated by Academia Sinica and NCHU. The major focus of my mentor’s lab is to understand how plants protect themselves against different heat stress conditions. I was given a project characterizing Arabidopsis mutants with defects in plant thermotolerance and cloning of the responsible genes. Fortunately, it became clear that my previous training in maize genetics made my life easier in mapping and cloning of the CLD1 gene, which, we later showed, encodes a long-sought enzyme that removes the phytol chain from chlorophyll during its turnover. I am very happy that the paper will be published in the same month of my wedding.

Current Position: Assistant Professor, Laboratory of Pomology, Graduate School of Agriculture, Kyoto University, Japan.
Education: PhD (2011), Graduate School of Agriculture, Kyoto University, Japan.
Non-scientific Interests: Orchestra performance (clarinet), piano, composition, fishing.
I have been associated with the field of Pomology, which is nested in Horticulture and studies on tree crops, since the beginning of my research life. In my Ph.D. studies, I investigated the astringency trait in persimmon fruit, mainly from the viewpoints of phytochemistry and molecular biology. After finishing my doctorate studies at Kyoto University, I was interested in learning new technologies, particularly bioinformatics. At just the right time, I fortunately had the opportunity to study as a visiting scientist with Drs. Luca Comai and Isabelle M. Henry at the Genome Center, University of California Davis. Using diploid Caucasian persimmon as the study material, we launched an investigation to identify dioecious sex determinant genes, which had not been isolated in any plant species. Our assembly of bioinformatics and evolutionary analyses rapidly enabled us to identify a Y-chromosome-encoded sex determinant, named OGI, and its counterpart gene, named MeGI. Continuing this excellent collaboration, we then tried to elucidate the variations in sexuality in various persimmons (Diospyros spp.), including the flexible sexuality in hexaploid Oriental persimmon (D. kaki), in the current study.
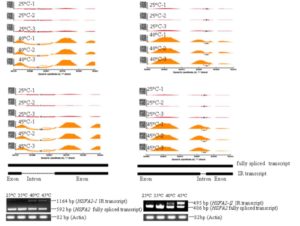 Heat stress is one of the main abiotic stresses plants encounter. Jiang et al. used combined transcriptomic and proteomic data to explore the responses of grape leaves to elevated temperatures (35, 40, 45°C). Using high-throughput sequencing and the iTRAQ (isobaric tags for relative and absolute quantitation) method, they found that non-random posttranscriptional events, particularly alternative splicing, are important heat responses. Notably, heat-shock proteins and transcription factors associated with high temperature were particularly prone to alternative splicing at elevated temperatures. How these differently-spliced transcripts affects plant heat response remains to be explored. Plant Physiol. 10.1104/pp.16.01305
Heat stress is one of the main abiotic stresses plants encounter. Jiang et al. used combined transcriptomic and proteomic data to explore the responses of grape leaves to elevated temperatures (35, 40, 45°C). Using high-throughput sequencing and the iTRAQ (isobaric tags for relative and absolute quantitation) method, they found that non-random posttranscriptional events, particularly alternative splicing, are important heat responses. Notably, heat-shock proteins and transcription factors associated with high temperature were particularly prone to alternative splicing at elevated temperatures. How these differently-spliced transcripts affects plant heat response remains to be explored. Plant Physiol. 10.1104/pp.16.01305 

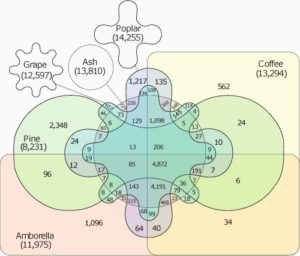 Ash dieback (a fungal disease) and the beetle Agrilus planipennis (a herbivore) are crushing ash tree populations in the Northern Hemisphere. To shed light on the genetic basis of the trees’ susceptibility and to understand the genetic diversity of these trees, Sollars et al. have sequenced one individual from the UK and annotated its genome. Through a deep sequence analysis, they compared the genome structure with other species for which genomes are already available, revealing an interesting set of genes shared among them. They also investigated the genetic diversity between different ash tree populations from Europe with the goal of finding the underlying genetic bases of tree susceptibility to fungal disease and herbivory. The authors found a correlation between ash dieback susceptibility and glycosides production (which defends against herbivory), indicating that it may be difficult to generate trees resistance to both fungal and herbivore attack. (Summary by
Ash dieback (a fungal disease) and the beetle Agrilus planipennis (a herbivore) are crushing ash tree populations in the Northern Hemisphere. To shed light on the genetic basis of the trees’ susceptibility and to understand the genetic diversity of these trees, Sollars et al. have sequenced one individual from the UK and annotated its genome. Through a deep sequence analysis, they compared the genome structure with other species for which genomes are already available, revealing an interesting set of genes shared among them. They also investigated the genetic diversity between different ash tree populations from Europe with the goal of finding the underlying genetic bases of tree susceptibility to fungal disease and herbivory. The authors found a correlation between ash dieback susceptibility and glycosides production (which defends against herbivory), indicating that it may be difficult to generate trees resistance to both fungal and herbivore attack. (Summary by 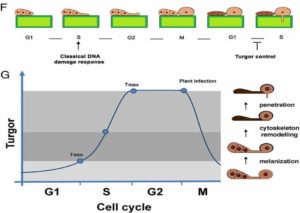 The rice blast fungus Magnaporthe oryzae causes a devastating disease of rice that can reduce harvests by 30%. Infection of plant tissues by the fungus requires the formation of an appressorium, a specialized structure that builds up sufficient pressure to burst through the plant cuticle. Previous work showed that cell cycle progression is required for appressorium formation and function.
The rice blast fungus Magnaporthe oryzae causes a devastating disease of rice that can reduce harvests by 30%. Infection of plant tissues by the fungus requires the formation of an appressorium, a specialized structure that builds up sufficient pressure to burst through the plant cuticle. Previous work showed that cell cycle progression is required for appressorium formation and function. 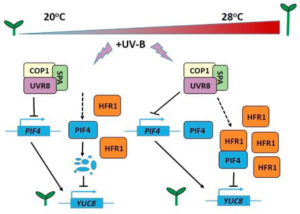 Ambient temperature can influence plant architecture, an effect termed thermomorphogenesis. In A. thaliana, thermomorphogenesis phenotypes include stem elongation and changes in leaf elevation angles. Increased auxin biosynthesis involving the transcription factor PIF4 is required for thermomorphogenesis, but the roles of other signals besides auxin remain unknown. Hayes et al. studied how ultraviolet-B (UV-B) light perceived through the photoreceptor UVR8 influences thermomorphogenesis using duplicate experiments at 20°C & 28°C. At 20°C & 28°C, PIF4 transcript abundance and thermomorphogenesis are both inhibited by signaling from heterodimers of UVR8 and the E3 ubiquitin ligase COP1. This is consistent with the decrease in PIF4 protein levels seen at 20°C, but there is no similar decrease for 28°C. At 28°C, the PIF4 protein is postulated to lose activity, and this loss is attributed to PIF4 binding to the bHLH protein HFR1 to form heterodimers that cannot bind DNA. UV-B light uses multiple mechanisms to inhibit thermomorphogenesis, and this explains one facet of how a plant interacts with and adapts to its environment. (Summary by
Ambient temperature can influence plant architecture, an effect termed thermomorphogenesis. In A. thaliana, thermomorphogenesis phenotypes include stem elongation and changes in leaf elevation angles. Increased auxin biosynthesis involving the transcription factor PIF4 is required for thermomorphogenesis, but the roles of other signals besides auxin remain unknown. Hayes et al. studied how ultraviolet-B (UV-B) light perceived through the photoreceptor UVR8 influences thermomorphogenesis using duplicate experiments at 20°C & 28°C. At 20°C & 28°C, PIF4 transcript abundance and thermomorphogenesis are both inhibited by signaling from heterodimers of UVR8 and the E3 ubiquitin ligase COP1. This is consistent with the decrease in PIF4 protein levels seen at 20°C, but there is no similar decrease for 28°C. At 28°C, the PIF4 protein is postulated to lose activity, and this loss is attributed to PIF4 binding to the bHLH protein HFR1 to form heterodimers that cannot bind DNA. UV-B light uses multiple mechanisms to inhibit thermomorphogenesis, and this explains one facet of how a plant interacts with and adapts to its environment. (Summary by 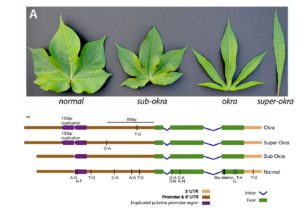 Homeodomain transcription factors are well-known as regulators of developmental patterning, including in leaves. Andres et al. examine the molecular basis behind leaf shape in cotton, particularly the Okra locus that was identified by breeders as a regulator of leaf shape. They show that the Okra locus encodes a homeodomain transcription factor, and that different leaf shapes (including normal, okra, super-okra and sub-okra) express allelic variants. Specifically, the narrower okra and super-okra leaves express higher levels of the gene. Suppressing this expression through viral-induced gene silencing transiently switches leaf shape to the broader normal form, demonstrating the importance of this gene in the control of leaf shape. As leaf shape is correlated with water-use efficiency, light interception and susceptibility to insects, these insights provide breeders with new tools. This paper would be a nice addition to a genetics course, especially if provided to students along with some of the classic studies of the Okra locus, such as
Homeodomain transcription factors are well-known as regulators of developmental patterning, including in leaves. Andres et al. examine the molecular basis behind leaf shape in cotton, particularly the Okra locus that was identified by breeders as a regulator of leaf shape. They show that the Okra locus encodes a homeodomain transcription factor, and that different leaf shapes (including normal, okra, super-okra and sub-okra) express allelic variants. Specifically, the narrower okra and super-okra leaves express higher levels of the gene. Suppressing this expression through viral-induced gene silencing transiently switches leaf shape to the broader normal form, demonstrating the importance of this gene in the control of leaf shape. As leaf shape is correlated with water-use efficiency, light interception and susceptibility to insects, these insights provide breeders with new tools. This paper would be a nice addition to a genetics course, especially if provided to students along with some of the classic studies of the Okra locus, such as 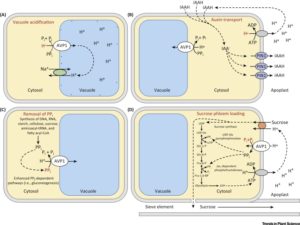 The AVP1 gene encodes a proton-pumping pyrophosphatase (H+-PPase) localized to the vacuolar membrane, which means that it pumps H+ into the vacuole using energy stored in pyrophosphatase (PPi). The direct consequences of its action are the acidification of the vacuole and the removal of PPi from the cytosol. Indirect consequences derived from these direct effects include enhanced salinity tolerance, due to sequestration of Na+ in the vacuole via Na+/H+ exchange. However, other effects, such as enhanced plant size in non-stressed conditions, are not so easy to explain. Schilling et al. review the diverse roles of AVP1, including effects on sucrose transport, inferred from knock-out and overexpression studies, and discuss how this gene can be used to increase crop yields. Trends Plant Sci.
The AVP1 gene encodes a proton-pumping pyrophosphatase (H+-PPase) localized to the vacuolar membrane, which means that it pumps H+ into the vacuole using energy stored in pyrophosphatase (PPi). The direct consequences of its action are the acidification of the vacuole and the removal of PPi from the cytosol. Indirect consequences derived from these direct effects include enhanced salinity tolerance, due to sequestration of Na+ in the vacuole via Na+/H+ exchange. However, other effects, such as enhanced plant size in non-stressed conditions, are not so easy to explain. Schilling et al. review the diverse roles of AVP1, including effects on sucrose transport, inferred from knock-out and overexpression studies, and discuss how this gene can be used to increase crop yields. Trends Plant Sci. 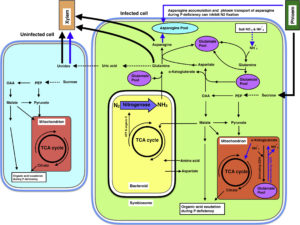 Legume nodules fix N, but their function has a high requirement for P, making nitrogen-fixation highly sensitive to P deficiency. Valentine et al. review how P limitation affects nodule function and also how nodules respond and adapt to P deficiency, drawing largely on studies of Virgilia divaricata, a legume endemic to P-poor soils of South Africa. One way that P deficiency affects nodule function is through the ADP:ATP ratio; N fixation and export from nodules has a high demand for energy. Although nodules are strong sinks for P, with P limitation NO3– and NH4+ can become preferred N sources (over N2), a switch that impacts amino acid and carbon skeleton availability. Lessons from this tropical legume can point to potential traits for legume crop improvement in other nutrient-poor ecosystems. Plant Science
Legume nodules fix N, but their function has a high requirement for P, making nitrogen-fixation highly sensitive to P deficiency. Valentine et al. review how P limitation affects nodule function and also how nodules respond and adapt to P deficiency, drawing largely on studies of Virgilia divaricata, a legume endemic to P-poor soils of South Africa. One way that P deficiency affects nodule function is through the ADP:ATP ratio; N fixation and export from nodules has a high demand for energy. Although nodules are strong sinks for P, with P limitation NO3– and NH4+ can become preferred N sources (over N2), a switch that impacts amino acid and carbon skeleton availability. Lessons from this tropical legume can point to potential traits for legume crop improvement in other nutrient-poor ecosystems. Plant Science 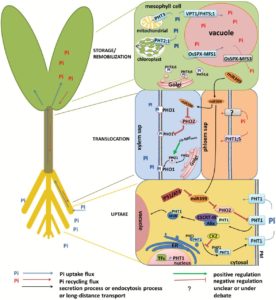 Nitrogen (N), phosphorus (P), and potassium (K) are the three macronutrients required in highest amounts for plant growth. N is abundant in the atmosphere, therefore plentiful if we overlook the energetic costs of converting N2 to usable form. By contrast, K and P are present in limited amounts in the Earth’s crust, and their extraction can be challenging. Over the next few decades, achieving good crop yields by applying these macronutrients to soils will become more and more expensive. Luan et al. review current understanding of K and P transporters and the regulatory circuits that control their activities. Ultimately, exploiting this knowledge should lead to greater uptake and use efficiencies of these essential nutrients. J. Exp. Bot.
Nitrogen (N), phosphorus (P), and potassium (K) are the three macronutrients required in highest amounts for plant growth. N is abundant in the atmosphere, therefore plentiful if we overlook the energetic costs of converting N2 to usable form. By contrast, K and P are present in limited amounts in the Earth’s crust, and their extraction can be challenging. Over the next few decades, achieving good crop yields by applying these macronutrients to soils will become more and more expensive. Luan et al. review current understanding of K and P transporters and the regulatory circuits that control their activities. Ultimately, exploiting this knowledge should lead to greater uptake and use efficiencies of these essential nutrients. J. Exp. Bot.  Current Position: Postdoctoral fellow in Agricultural Biotechnology Research Center, Academia Sinica, Taiwan.
Current Position: Postdoctoral fellow in Agricultural Biotechnology Research Center, Academia Sinica, Taiwan.