New kid on the plant block: Single-cell proteomics
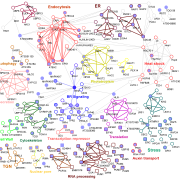
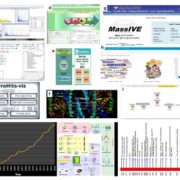
 While single-cell omics technologies, particularly transcriptomics, are already becoming widely adopted in plant science, quantifying proteins at single cell resolution is less established. Fortunately, important technological strides have been made that improve sample preparation, separation techniques, and overall sensitivity and resolution to make single cell proteomics (SCP) possible. Montes et al. have recently developed a multiplexed methodology for SCP specifically adapted to plant tissue, overcoming typical challenges associated with handling such samples. As a proof-of-concept, the authors compared proteins from two adjacent root cell layers, the endodermis and cortex, and observed their distinct proteomic signatures. The optimized SCP can successfully detect proteins associated with both low and high abundance transcripts, and even detects low abundant proteins like transcription factors, opening up tremendous possibilities to study plant signaling networks in neighboring cell types. As expected however, the varying cell sizes along the root led to considerable variation in protein quantification, an important consideration for improvement in future studies. In conclusion, this methodology offers a clear, comprehensive, and practical tool for scientists looking to explore the world of single-cell proteomics in plants. (Summary by Thomas Depaepe @thdpaepe). New Phytologist 10.1111/nph.19923
While single-cell omics technologies, particularly transcriptomics, are already becoming widely adopted in plant science, quantifying proteins at single cell resolution is less established. Fortunately, important technological strides have been made that improve sample preparation, separation techniques, and overall sensitivity and resolution to make single cell proteomics (SCP) possible. Montes et al. have recently developed a multiplexed methodology for SCP specifically adapted to plant tissue, overcoming typical challenges associated with handling such samples. As a proof-of-concept, the authors compared proteins from two adjacent root cell layers, the endodermis and cortex, and observed their distinct proteomic signatures. The optimized SCP can successfully detect proteins associated with both low and high abundance transcripts, and even detects low abundant proteins like transcription factors, opening up tremendous possibilities to study plant signaling networks in neighboring cell types. As expected however, the varying cell sizes along the root led to considerable variation in protein quantification, an important consideration for improvement in future studies. In conclusion, this methodology offers a clear, comprehensive, and practical tool for scientists looking to explore the world of single-cell proteomics in plants. (Summary by Thomas Depaepe @thdpaepe). New Phytologist 10.1111/nph.19923