Multidimensional gene regulatory landscape of a bacterial pathogen in plants (Nature Plants)
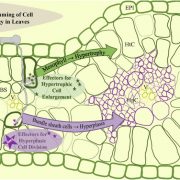
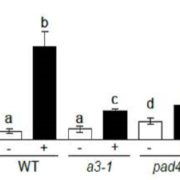
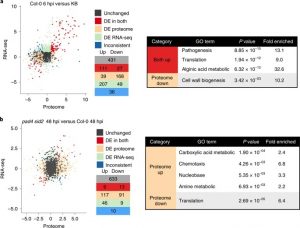 The outcome of a plant-bacteria interaction is determined by the bacterial virulence and plant immune systems. Individually, these systems, and to an extent their interactions, have been well studied. However, certain aspects of their interactions remain elusive, particularly how plant immunity affects bacterial functions. Nobori et al. used an integrated approach of transcriptomics and proteomics analyses to understand how plant immunity modulates bacteria mRNA and protein expression in the Arabidopsis thaliana – Pseudomonas syringae pathosystem. Virulent and an avirulent strains of P. syringae were used. In order to understand the role of the SA pathway and effector-triggered immunity (ETI) on bacterial processes, compromised mutants were included in the analysis. Samples were collected at 6- and 48-hours post infection in order to understand the dynamic changes in mRNA and protein expression at the early and late stage of infection. Comparative analysis showed that mRNA expression moderately correlates with protein expression in all conditions, suggesting that mRNA expression closely reflects protein expression in P. syringae. Further analysis also showed that changes in the bacterial transcriptome are good predictors of changes in the proteome during plant colonization. However, there are a few cases in which mRNA expression changes did not correlate with protein expression; notably, the bacterial cell wall biogenesis-related function was suppressed at the protein but not mRNA level. Finally, previously unknown gene regulatory modules that are important for virulence were also identified. (Summary by Toluwase Olukayode) @toluxylic Nature Plants 10.1038/s41477-020-0690-7
The outcome of a plant-bacteria interaction is determined by the bacterial virulence and plant immune systems. Individually, these systems, and to an extent their interactions, have been well studied. However, certain aspects of their interactions remain elusive, particularly how plant immunity affects bacterial functions. Nobori et al. used an integrated approach of transcriptomics and proteomics analyses to understand how plant immunity modulates bacteria mRNA and protein expression in the Arabidopsis thaliana – Pseudomonas syringae pathosystem. Virulent and an avirulent strains of P. syringae were used. In order to understand the role of the SA pathway and effector-triggered immunity (ETI) on bacterial processes, compromised mutants were included in the analysis. Samples were collected at 6- and 48-hours post infection in order to understand the dynamic changes in mRNA and protein expression at the early and late stage of infection. Comparative analysis showed that mRNA expression moderately correlates with protein expression in all conditions, suggesting that mRNA expression closely reflects protein expression in P. syringae. Further analysis also showed that changes in the bacterial transcriptome are good predictors of changes in the proteome during plant colonization. However, there are a few cases in which mRNA expression changes did not correlate with protein expression; notably, the bacterial cell wall biogenesis-related function was suppressed at the protein but not mRNA level. Finally, previously unknown gene regulatory modules that are important for virulence were also identified. (Summary by Toluwase Olukayode) @toluxylic Nature Plants 10.1038/s41477-020-0690-7