More complexity for root meristem development
Xiong et al. investigated the transcriptional regulation of PLT1 in Arabidopsis, which revolves around competitive and antagonistic binding of JANUS and GIF1 to IMPORTIN β4. The Plant Cell (2020). https://doi.org/10.1105/tpc.20.00108
By Feng Xiong and Sha Li, State Key Laboratory of Crop Biology, College of Life Sciences, Shandong Agricultural University
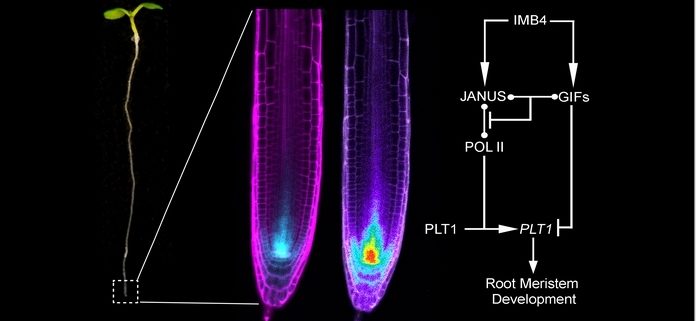
Background: Spatially coordinated cell division and differentiation during the development of the root meristem (RAM) are critical steps for sustained root growth in plants. Recent studies demonstrated that the transcription factor PLETHORA1 (PLT1) forms a concentration gradient to control RAM development. However, how the transcriptional regulation of PLT1 contributes to its protein gradient remains unknown. Two Arabidopsis proteins we have been working on, JANUS and IMPORTIN β4 (IMB4), play positive yet unclear roles in root growth. JANUS is a putative spliceosome subunit with transcriptional activity through its binding to RNA polymerase II (Pol II), while IMB4 is involved in protein trafficking between the nucleus and the cytoplasm or in maintaining protein stability.
Question: We aimed to identify the transcriptional pathways that act upstream of PLT1 and understand the complexity of the underlying regulatory networks controlling RAM development.
Findings: We discovered that Arabidopsis PLT1 activates its own transcription and that this auto-transcriptional regulation relies on JANUS. JANUS interacts with PLT1 and Pol II through different domains to facilitate PLT1 transcription. The JANUS-dependent PLT1 auto-transcriptional activation potentially amplifies the PLT1 gradient established by mitotic dilution and intercellular movement: it is thus critical for the maintenance of sufficiently high levels of PLT1 during RAM development. By contrast, a transcriptional co-factor, GRF-INTERACTING FACTOR1/ANGUSTIFOLIA3 (GIF1/AN3), interacts with JANUS to deplete it from the JANUS-Pol II transcriptional complex, therefore negatively regulating JANUS-dependent PLT1 transcription. As an additional layer of complexity, GIF1/AN3 and JANUS both rely on IMB4 for their nuclear accumulation.
Next steps: That two interacting but antagonistic regulators of PLT1 are regulated by the same importin raises the following question: how is selectivity of interaction between IMB4, JANUS, and GIF1/AN3 spatially controlled in the context of root development? Plant genomes encode few importins, even though the demand for nucleo-cytoplasmic shuttling is tremendous. Whether one importin is truly responsible for various cargos in the same signaling pathway for logistical benefits is certainly worthy of future investigation.