Modulation of gene expression in plants with orthogonal regulatory systems (Nat. Chem. Biol.)
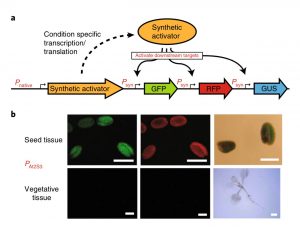 Some of the most promising applications in synthetic biology need precise control of gene expression. For instance, metabolic engineering in plants requires the expression of enzyme-coding genes at a precise time, space, and quantity to ensure correct output. Recently, Belcher, Vuu, and colleagues engineered an orthogonal system (that does not interfere with other cellular processes) to modulate gene expression in plants. They exploited the well-characterized yeast GAL4 transcriptional module and adapted it for use in plants. They characterized the system at two primary levels: cis-elements, sequences in the promoters; and trans-elements, modifications in activations/repression domains in the transcription factors (TF). The authors tested promoter strength using combinations of different cis-elements from yeast and minimal promoters from plants. To develop systems where gene expression can be induced in specific tissues or in response to specific stimuli and with controlled levels of activity, the authors used the tissue-specific activators (At2S3, endosperm-specific), or environmental response activators (AtPht1.1, low phosphate-induced), and explored other transcription factors such as MADS, HOMEOBOX, GATA, and bZIP as additional trans-elements, which allowed them to develop hybrid promoters designed to bind multiple TFs to have more precise control of the transcriptional output. This work opens the door to introducing more complex genetic circuits to plants, and will allow the generation of new tools to modulate gene expression in plants for synthetic biology applications. (Summary by Humberto Herrera Ubaldo @herrera_h) Nature Chem. Biol. 10.1038/s41589-020-0547-4
Some of the most promising applications in synthetic biology need precise control of gene expression. For instance, metabolic engineering in plants requires the expression of enzyme-coding genes at a precise time, space, and quantity to ensure correct output. Recently, Belcher, Vuu, and colleagues engineered an orthogonal system (that does not interfere with other cellular processes) to modulate gene expression in plants. They exploited the well-characterized yeast GAL4 transcriptional module and adapted it for use in plants. They characterized the system at two primary levels: cis-elements, sequences in the promoters; and trans-elements, modifications in activations/repression domains in the transcription factors (TF). The authors tested promoter strength using combinations of different cis-elements from yeast and minimal promoters from plants. To develop systems where gene expression can be induced in specific tissues or in response to specific stimuli and with controlled levels of activity, the authors used the tissue-specific activators (At2S3, endosperm-specific), or environmental response activators (AtPht1.1, low phosphate-induced), and explored other transcription factors such as MADS, HOMEOBOX, GATA, and bZIP as additional trans-elements, which allowed them to develop hybrid promoters designed to bind multiple TFs to have more precise control of the transcriptional output. This work opens the door to introducing more complex genetic circuits to plants, and will allow the generation of new tools to modulate gene expression in plants for synthetic biology applications. (Summary by Humberto Herrera Ubaldo @herrera_h) Nature Chem. Biol. 10.1038/s41589-020-0547-4