Mightier than Muscle: A Near-atomic View of Pollen Actin Filaments
Beyond its well-known role in muscle contraction, the cytoskeletal component actin participates in many critical cellular processes. Globular actin polymerizes into thin, flexible filaments that assemble into dynamic higher-order structures, such as bundles and networks. In addition to providing mechanical support, these structures organize the contents of the cytoplasm and play fundamental roles in cell motility, vesicle trafficking, and cell signaling (Szymanski and Staiger, 2018). An array of actin-binding proteins regulates the polymerization and depolymerization of actin and directs its organization into higher-order structures (Winder and Ayscough, 2005). While the cellular functions and biochemical activities of actin are well-established in plants, the structural basis for these properties is unclear.
 Now, in a Breakthrough report, Ren and colleagues (2019) present the first near-atomic structure of plant actin filaments. Using cryogenic electron microscopy (cryo-EM) and a real-space helical reconstruction approach, the authors determined the structure of Zea mays (maize) pollen actin (ZMPA) filaments, polymerized in vitro, at a resolution of 3.9 Å. They compared this structure with published structures of rabbit skeletal muscle actin (RSMA; Galkin et al., 2015; von der Ecken et al., 2015; Merino et al., 2018) and jasplakinolide-stabilized Plasmodium falciparum (malaria parasite) actin (JASP-PfAct1; Pospich et al., 2017) filaments.
Now, in a Breakthrough report, Ren and colleagues (2019) present the first near-atomic structure of plant actin filaments. Using cryogenic electron microscopy (cryo-EM) and a real-space helical reconstruction approach, the authors determined the structure of Zea mays (maize) pollen actin (ZMPA) filaments, polymerized in vitro, at a resolution of 3.9 Å. They compared this structure with published structures of rabbit skeletal muscle actin (RSMA; Galkin et al., 2015; von der Ecken et al., 2015; Merino et al., 2018) and jasplakinolide-stabilized Plasmodium falciparum (malaria parasite) actin (JASP-PfAct1; Pospich et al., 2017) filaments.
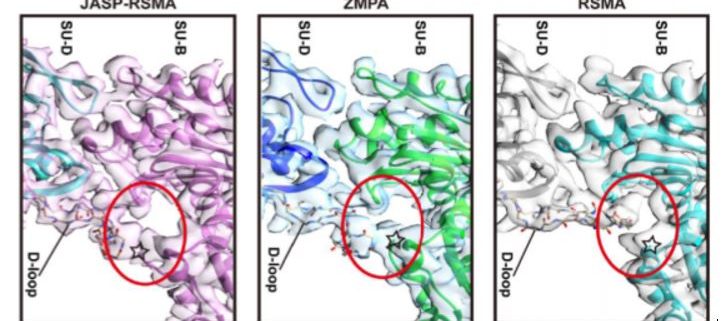
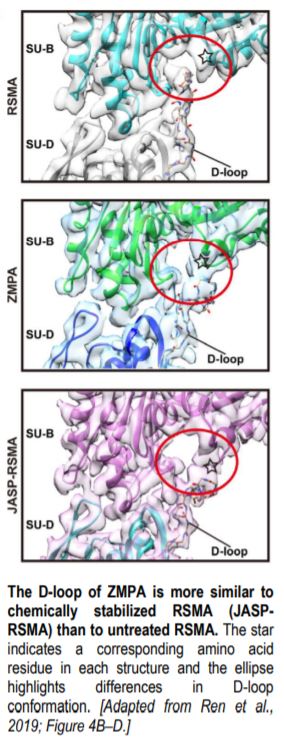
The overall structure of ZMPA filaments resembles those of RSMA and JASP-PfAct1. ZMPA filaments can be described as a double-stranded helix, with a helical twist of –166.77° and a helical rise of 27.5 Å. Each ZMPA subunit has four subdomains (SD1–4). SD2 contains a D-loop, a conserved region implicated in actin polymerization and stability (Grintsevich et al., 2017). The staggered actin subunits within the filament are stabilized by various intra- and interstrand interactions; the authors mapped the interaction sites and noted that several were conserved in RSMA (von der Ecken, 2015) and JASP-PfAct1 (Pospich et al., 2017) filaments.
Intriguingly, the D-loop of ZMPA differs from that of RSMA, but resembles the D-loop of RSMA that has been chemically stabilized by jasplakinolide (JASP-RMSA, figure), suggesting that ZMPA filaments are more stable than their mammalian counterparts. Furthermore, single-molecule magnetic tweezers analysis showed that ZMPA can resist a greater stretching force than RSMA.
The finding that actin filaments from pollen are more resistant to stretching than those from muscle was unexpected. Pollen tubes are remarkable tip-growing cells, capable of extending many thousands of times their width, as they shuttle sperm cells through the style to the ovule. In contrast to the parallel arrangement of actin filaments in the contractile unit of muscle cells, growing pollen tubes are characterized by long bundles of actin in the shank and a fringe of actin near the apex. The actin cytoskeleton in the apex of growing pollen tubes is more dynamic than that in mature muscle cells; indeed, pollen tube growth depends on actin polymerization (Vidali and Hepler, 2001). It would be fascinating to determine whether the conformation of the D-loop reported here is unique to tip-growing cells or is a consistent feature of plant cell actin filaments and to investigate the effect of structural features such as the D-loop on cellular function and biochemical activity. This work sets the stage for such studies.
Kathleen L. Farquharson
Science Editor
http://orcid.org/0000-0002-8032-0041
References
Galkin, V.E., Orlova, A., Vos, M.R., Schroder, G.F., and Egelman, E.H. (2015). Near-atomic resolution for one state of F-actin. Structure 23: 173-182.
Grintsevich, E.E., Ge, P., Sawaya, M.R., Yesilyurt, H.G., Terman, J.R., Zhou, Z.H., and Reisler, E. (2017). Catastrophic disassembly of actin filaments via Mical-mediated oxidation. Nat. Commun. 8: 2183.
Merino, F., Pospich, S., Funk, J., Wagner, T., Kullmer, F., Arndt, H.D., Bieling, P., and Raunser, S. (2018). Structural transitions of F-actin upon ATP hydrolysis at near-atomic resolution revealed by cryo-EM. Nat. Struct. Mol. Biol. 25: 528-537.
Pospich, S., Kumpula, E.P., von der Ecken, J., Vahokoski, J., Kursula, I., and Raunser, S. (2017). Near-atomic structure of jasplakinolide-stabilized malaria parasite F-actin reveals the structural basis of filament instability. Proc. Natl. Acad. Sci. USA 114: 10636-10641.
Ren, Z., Zhang, Y., Zhang, Y., He, Y., Du, P., Wang, Z., Sun, F., and Ren. H. Cryo-EM Structure of Actin Filaments from Zea mays Pollen. 2019. Plant Cell https://doi.org/10.1105/tpc.19.00973.
Szymanski D. and Staiger C.J. (2018.) The actin cytoskeleton: functional arrays for cytoplasmic organization and cell shape control. Plant Physiol. 176(1):106–118. doi:10.1104/pp.17.01519.
Winder, S.J. and Ayscough, K.R. (2005). Actin-binding proteins. Journal of Cell Science 2005 118: 651-654.
Vidali, L. and Hepler, P.K. Actin and pollen tube growth. Protoplasma (2001) 215: 64–76.
Von der Ecken, J., Muller, M., Lehman, W., Manstein, D.J., Penczek, P.A., and Raunser, S. (2015). Structure of the F-actin-tropomyosin complex. Nature 519: 114-117.