Micro Manager: MicroRNA Dynamics Facilitate Correct Embryo Morphogenesis and Patterning
MicroRNAs (miRNAs) are a class of small (~21 nucleotide) non-coding RNAs that function as post-transcriptional repressors of gene expression. In plants, miRNAs recognize their target mRNAs based on perfect, or near-perfect, sequence complementarity, ultimately mediating their cleavage and/or translational inhibition (Jones-Rhoades et al., 2006). Most miRNA targets identified to date encode transcription factors (TFs); miRNA-TF interactions have been described for all major TF families known to be involved in plant development and environmental stress responses. Hence, miRNAs function as key regulators of multiple aspects of plant physiology, either directly or indirectly through regulatory cascades.
Several studies have highlighted the importance of particular miRNAs in cellular differentiation and developmental timing in post-embryonic tissues. However, far less is known about their involvement in the earlier patterning events that occur during embryogenesis and are crucial for establishing the basic body plan of the plant. Here, Plotnikova et al. (2019) describe the profiling and functional characterization of Arabidopsis thaliana embryonic miRNA populations expressed in eight different developmental stages spanning the pre-cotyledon, transition, and maturation phases of embryogenesis.
In total, 349 miRNAs from 259 families were identified using a novel high-throughput sequencing technique specifically developed to handle low amounts of input RNA whilst simultaneously enriching for small RNAs. Many of these miRNAs were differentially expressed across the high-resolution dataset, enabling miRNAs to be clustered according to their temporal expression dynamics. Furthermore, whole-mount RNA in situ hybridizations revealed that miRNA families exhibit distinctive spatial expression patterns across various cell types in the developing embryo. Taken together, this suggests that groups of miRNAs function cooperatively during the various phases of embryogenesis to restrict target gene expression spatially and/or temporally.
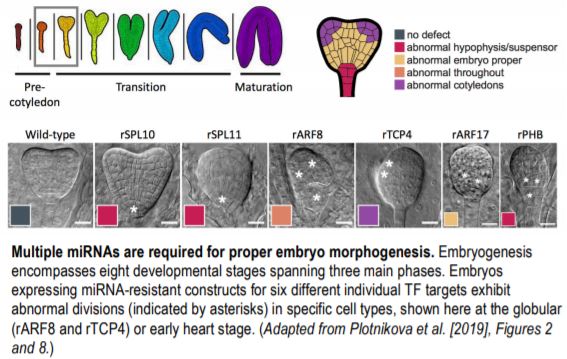 To determine the likely targets of the embryonic miRNAs, the authors utilized their previously developed nanoPARE next-generation sequencing methodology, identifying 59 high-confidence transcript RNA cleavage sites, 30 of which encode TFs. Six of these TFs (SPL10/11, ARF17, PHB, ARF8, and TCP4) were then examined in more detail at both the cellular and whole embryo level. A GFP-based sensor system was designed to detect miRNA-mediated degradation of the individual TF target sequences. The spatial dynamics of TF repression across the different embryo cell types aligned with those described from in situ analysis of the analogous miRNA, with some miRNAs exhibiting an even broader repression domain. Accordingly, embryos from transgenic plant lines expressing constructs with synonymous mutations in the miRNA target sequences, rendering them ‘miRNA-resistant’, exhibited clear morphological defects. Crucially, the observed phenotypes were consistent with abnormal cellular divisions occurring in the cell types and/or embryo stage in which the corresponding miRNAs were active.
To determine the likely targets of the embryonic miRNAs, the authors utilized their previously developed nanoPARE next-generation sequencing methodology, identifying 59 high-confidence transcript RNA cleavage sites, 30 of which encode TFs. Six of these TFs (SPL10/11, ARF17, PHB, ARF8, and TCP4) were then examined in more detail at both the cellular and whole embryo level. A GFP-based sensor system was designed to detect miRNA-mediated degradation of the individual TF target sequences. The spatial dynamics of TF repression across the different embryo cell types aligned with those described from in situ analysis of the analogous miRNA, with some miRNAs exhibiting an even broader repression domain. Accordingly, embryos from transgenic plant lines expressing constructs with synonymous mutations in the miRNA target sequences, rendering them ‘miRNA-resistant’, exhibited clear morphological defects. Crucially, the observed phenotypes were consistent with abnormal cellular divisions occurring in the cell types and/or embryo stage in which the corresponding miRNAs were active.
In plants, primary miRNA transcripts are processed into mature miRNAs by DICER-LIKE1 (DCL1). Unsurprisingly, dcl1-null mutants exhibit abnormal cellular divisions, arresting early in embryo development. Therefore, dcl1 embryos are a useful tool for further deciphering the roles of miRNAs during embryogenesis. Here, the authors utilized dcl1 as both a negative control in their analysis of the spatiotemporal dynamics of miRNA/TF target expression and to confirm the validity of the identified TF target cleavage products which, as expected, were absent in the mutant. Indeed, the global dcl1 embryo transcriptome is significantly altered from that of the equivalent wild type, with ectopic, precocious, and premature expression of multiple late-stage transcripts. This finding, together with other results of the study, provides strong evidence for the involvement and crosstalk of multiple miRNA-regulated transcriptional cascades during embryogenesis.
This detailed transcriptional analysis of miRNA expression dynamics and functionality across embryogenesis, together with the associated transcriptome dataset of coding RNAs (Hofmann et al., 2019), provides novel insights into the molecular mechanisms underpinning early plant patterning events. Since individual miRNAs can have multiple mRNA targets, one future challenge will be to elucidate miRNA interactions, and the roles of other small RNAs, in the whole embryogenesis gene regulatory network.
Emily Breeze
School of Life Sciences,
University of Warwick, Coventry, UK
ORCID ID: 0000-0001-5383-5448
REFERENCES
Hofmann, F., Schon, M.A., and Nodine, M.D. (2019). The embryonic transcriptome of Arabidopsis thaliana. Plant Reproduction 32: 77–91.
Jones-Rhoades, M.W., Bartel, D.P., and Bartel, B. (2006). MicroRNAS and their regulatory roles in plants. Ann. Rev. Plant Biol. 57: 19–53.
Plotnikova, A., Kellner, M.J., Schon, M.A., Mosiolek, M., and Nodine, M.D. (2019). MicroRNA Dynamics and Functions During Arabidopsis Embryogenesis. Plant Cell 31: https://doi.org/10.1015/tpc.19.00395




