MASS proteins and stomatal development in Arabidopsis (PLOS Genetics)
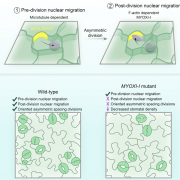
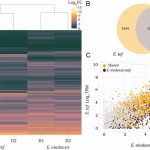
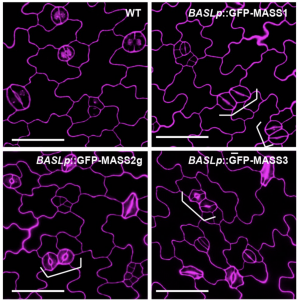 Stomata are the pores surrounded by a pair of guard cells on the plant epidermis that help in gaseous exchange. The number and spacing of stomata are regulated by a series of phosphorylation and de-phosphorylation events of key transcription factors through the mitogen-activated protein kinase (MAPK) signal cascade [which includes MAPK kinases (MAPKKs), and MAPKK kinases (MAPKKKs)]. One of the well-known pathways in stomatal development is a series of phosphorylation events by YODA (a MAPKKK), MAPKK4/5, and MAPK 3/6, leading to the eventual inhibition of SPEECHLESS, transcription factor involved in stomatal development. In this paper Xue et al. characterized three other substrates of MAPK signaling that they named MAPK SUBSTRATES IN THE STOMATAL LINEAGE (MASS) proteins. Using biochemical and genetic experiments, the authors showed that MASS proteins regulate the number and spacing between adjacent stomata. Further, mechanistically MASS proteins are localized to the plasma membrane and interact with YODA to negatively regulate the MAPK signaling. Thus, this paper advances our understanding of stomatal development by adding three more MASS proteins in the stomatal developmental pathway. (Summary by Vijaya Batthula @Vijaya_Batthula). PLOS Genetics 10.1371/journal.pgen.1008706
Stomata are the pores surrounded by a pair of guard cells on the plant epidermis that help in gaseous exchange. The number and spacing of stomata are regulated by a series of phosphorylation and de-phosphorylation events of key transcription factors through the mitogen-activated protein kinase (MAPK) signal cascade [which includes MAPK kinases (MAPKKs), and MAPKK kinases (MAPKKKs)]. One of the well-known pathways in stomatal development is a series of phosphorylation events by YODA (a MAPKKK), MAPKK4/5, and MAPK 3/6, leading to the eventual inhibition of SPEECHLESS, transcription factor involved in stomatal development. In this paper Xue et al. characterized three other substrates of MAPK signaling that they named MAPK SUBSTRATES IN THE STOMATAL LINEAGE (MASS) proteins. Using biochemical and genetic experiments, the authors showed that MASS proteins regulate the number and spacing between adjacent stomata. Further, mechanistically MASS proteins are localized to the plasma membrane and interact with YODA to negatively regulate the MAPK signaling. Thus, this paper advances our understanding of stomatal development by adding three more MASS proteins in the stomatal developmental pathway. (Summary by Vijaya Batthula @Vijaya_Batthula). PLOS Genetics 10.1371/journal.pgen.1008706
[altmetric doi=”10.1371/journal.pgen.1008706″ details=”right” float=”right”]