Lignification and oxidative enzymes: Localization, localization, localization!
Name Marc Somssich
Affiliation University of Melbourne
ORCiD 0000-0001-5092-6168
Lignification of cell walls is a pivotal process in plant life. Lignin is a biopolymer that provides additional stability to secondary cell walls (SCWs), enables vascular cells to transport water, and protects the plant from pathogenic attack (Tobimatsu and Schuetz, 2019). It is produced from monolignols which are synthesized in the cytosol and subsequently secreted to the apoplast, where they are cross-linked via radical–radical coupling. Oxidative enzymes, laccases (LAC) and peroxidases (PRX), as well as reactive oxygen species (ROS) such as H2O2, are required for this polymerization step. The polymerized lignin is incorporated into the SCW, where it cross-links the different cell wall polysaccharides. However, only certain cell types have lignified SCWs, and even in these specialized cells there can be local differences in lignification between the walls. Lignification therefore appears to be a tightly directed and precisely localized event. From recent work it has emerged that this precise targeting is in part achieved by specific localizations of the oxidative enzymes. In this regard, the lab of A. Lacey Samuels has previously described the localization of LAC4 and PRX64 to either the entire SCW (LAC4), or just to the middle lamella and cell corners (PRX64) (Schuetz et al., 2014; Yi Chou et al., 2018). Building on this work, they now set out to assign several more LAC and PRX enzymes involved in lignification to their specific sub-domain of the SCW, bringing us closer to visualizing lignin polymerization on a sub-cellular level (Hoffmann et al., 2020).
In their new manuscript in this issue of Plant Physiology, Hoffmann et al. use expression data for different LAC and PRX genes, data on co-expression of these genes with other SCW synthesis components, and known phenotypes of mutants, to narrow down the long list of Arabidopsis thaliana LAC and PRX family members to eight prime candidates, which might be the central enzymes facilitating lignification of xylem cells and interfascicular fibers in Arabidopsis stems. This thorough analysis forms the basis of the work presented here, and the authors have included a helpful table summarizing LAC and PRX gene IDs, expression patterns and co-expression data, as well as stem mutant phenotypes. A lot of the work on understanding the role of LAC in lignin polymerization has focused on LAC4 and LAC17, with LAC4 being the most highly expressed LAC in stems, and lac4 lac17 double mutants exhibiting up to 40% reduced lignification in stems (Berthet et al., 2011; Hall and Ellis, 2013). Similarly, work on PRX has often focused on PRX64, the most highly expressed PRX in stems (Hall and Ellis, 2013; Yi Chou et al., 2018).
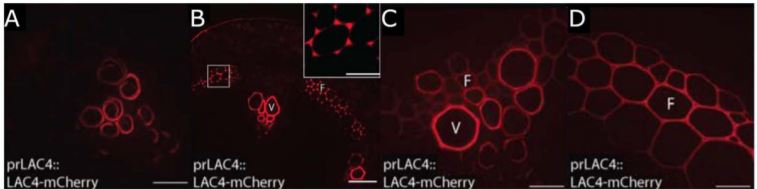
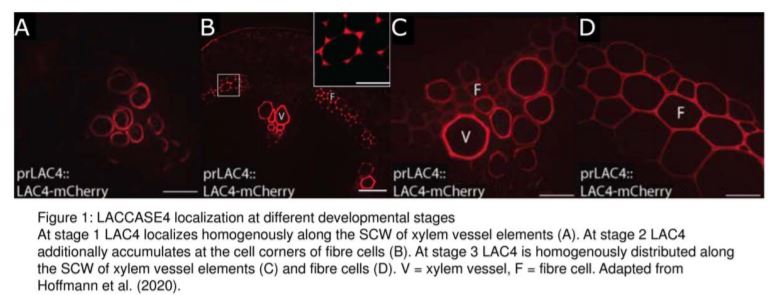 In their bioinformatic analysis, Hoffmann et al. additionally identified LAC10, PRX42, PRX52, PRX71, and PRX72 as the most promising candidates to study lignification in stems. Tagging these proteins with the mCherry fluorophore, they then visualized the localization of these eight enzymes in cells of the Arabidopsis stem, sub-domains of the SCW, as well as comparing several developmental stages. They found that LAC17 and PRX72 localize to the SCW of xylem vessels and fibre cells, while PRX64 and PRX71 localize to the cell corners and middle lamella of fibre cells. LAC10, PRX42, and PRX52 localize to non-lignified tissues, and may therefore be active in other pathways. Interestingly, LAC4 is distributed homogenously along the SCW of protoxylem vessel elements at the earliest developmental stages, and then additionally localizes specifically to the cell corners and middle lamella of fibre cells, before it also shows a homogenous distribution in the SCW of these cells for the remaining developmental stages (Fig. 1). LAC4 may therefore play a role in lignification of the SCW in both vessel elements and fibre cells during different stages of development.
In their bioinformatic analysis, Hoffmann et al. additionally identified LAC10, PRX42, PRX52, PRX71, and PRX72 as the most promising candidates to study lignification in stems. Tagging these proteins with the mCherry fluorophore, they then visualized the localization of these eight enzymes in cells of the Arabidopsis stem, sub-domains of the SCW, as well as comparing several developmental stages. They found that LAC17 and PRX72 localize to the SCW of xylem vessels and fibre cells, while PRX64 and PRX71 localize to the cell corners and middle lamella of fibre cells. LAC10, PRX42, and PRX52 localize to non-lignified tissues, and may therefore be active in other pathways. Interestingly, LAC4 is distributed homogenously along the SCW of protoxylem vessel elements at the earliest developmental stages, and then additionally localizes specifically to the cell corners and middle lamella of fibre cells, before it also shows a homogenous distribution in the SCW of these cells for the remaining developmental stages (Fig. 1). LAC4 may therefore play a role in lignification of the SCW in both vessel elements and fibre cells during different stages of development.
Having demonstrated these specific localizations for the different enzymes, the authors move on to test for actual oxidizing activity in these regions. Incubating stem sections with the PRX-oxidation substrate TMB (3,3’,5,5’-tetramethylbenzidine) and exogenous H2O2 results in oxidized TMB in all cell walls, independent of lignification, indicating that PRXs are generally present in all walls. When the stem sections were incubated with TMB alone, however, oxidation was limited to the lignified SCW, overlapping with the localization of the different LAC and PRX enzymes. This observation demonstrates that it is not just the localization of the enzymes to these specific regions, but also targeted release of endogenous H2O2 into the apoplast in these domains that contributes to the specific lignification pattern.
Hoffmann et al. have identified LAC and PRX enzymes (as well as combinations thereof) involved in lignification of SCW at different developmental stages and in specific sub-domains of the SCW, which will help to refine our model of lignification. They further demonstrate that it is not just the spatial distribution of the different enzyme sets and the targeted secretion of monolignols to these regions, but also localized ROS accumulation, that is required to achieve highly localized lignification. This is an agreement with recent findings that lignification of the Casparian strip is also dependent on targeted ROS accumulation and oxidative enzyme localization (Fujita et al., 2020).
The findings presented here also demonstrate that the many LAC and PRX family members are not merely redundant to each other, but appear to fulfill specific functions in specific domains and at specific time points. It will be interesting to learn more about how the enzymes are directed towards their specific domains, and how they are sequestered there. In this regard, LAC4, which changes its intramural localization from the corners of fiber cells, to a homogeneous distribution, will be an especially interesting candidate. Overall, the results in this publication represent another step towards understanding lignin polymerization at the sub-cellular level.
LITERATURE CITED
Berthet S, Demont-Caulet N, Pollet B, Bidzinski P, Cézard L, Le Bris P, Borrega N, Hervé J, Blondet E, Balzergue S, et al (2011) Disruption of LACCASE4 and 17 Results in Tissue-Specific Alterations to Lignification of Arabidopsis thaliana Stems. Plant Cell 23: 1124–1137
Fujita S, Bellis D De, Edel KH, Köster P, Andersen TG, Schmid-siegert E, Tendon VD, Pfister A, Marhavý P, Ursache R, et al (2020) SCHENGEN receptor module drives localized ROS production and lignification in plant roots. EMBO J 1–18
Hall H, Ellis B (2013) Transcriptional programming during cell wall maturation in the expanding Arabidopsis stem. BMC Plant Biol 13: 14
Schuetz M, Benske A, Smith RA, Watanabe Y, Tobimatsu Y, Ralph J, Demura T, Ellis B, Samuels AL (2014) Laccases direct lignification in the discrete secondary cell wall domains of protoxylem. Plant Physiol 166: 798–807
Tobimatsu Y, Schuetz M (2019) Lignin polymerization: how do plants manage the chemistry so well? Curr Opin Biotechnol 56: 75–81
Yi Chou E, Schuetz M, Hoffmann N, Watanabe Y, Sibout R, Samuels AL (2018) Distribution, mobility, and anchoring of lignin-related oxidative enzymes in Arabidopsis secondary cell walls. J Exp Bot 69: 1849–1859