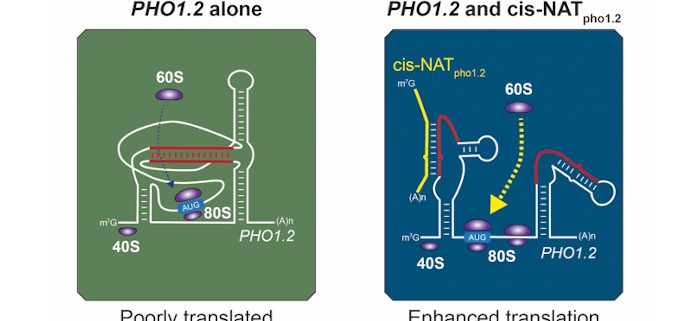
Key role of RNA structure in a sense-antisense RNA interaction and enhancement of translation
The Plant Cell
Rodrigo S. Reis1 and Yves Poirier1
1 Department of Plant Molecular Biology, University of Lausanne, Lausanne, Switzerland
Background: A large proportion of plant RNAs are natural antisense transcripts (NAT), non-coding RNAs that are complementary to sense, protein-coding, mRNAs. The co-expression of sense and antisense RNAs is thought to primarily result in the suppression of gene expression, because it is assumed that double-stranded RNAs (dsRNA) would form within their complementary sense-antisense sequences, which is a typical substrate for endoribonucleases, resulting in mRNA degradation and gene silencing. This assumption has recently been challenged by the discovery that the expression of some NATs in plants (e.g. at the rice PHO1.2 locus) and human actually enhances sense mRNA translation.
Question: We aimed to understand how the interaction between rice PHO1.2 mRNA (encoding a phosphate exporter) and its cognate NAT (cis-NATpho1.2) enhances PHO1.2 mRNA translation.
Findings: We first determined the structure of both the sense mRNA and NAT using high throughput RNA sequencing. This revealed that the RNA structure of PHO1.2 mRNA is altered at specific nucleotides in the presence of its cis-NATpho1.2. Similarly, specific and localised changes were found in cis-NATpho1.2 in the presence of PHO1.2 mRNA. This was in clear contrast with an expected widespread formation of dsRNA. Indeed, our data support a model in which these two RNAs maintain most of their intra-molecular secondary structures upon interaction, and only partially “unfold” at specific regions to form inter-molecular interactions. Subsequently, we identified the minimal region within cis-NATpho1.2 that is responsible for translation enhancement, as well as a specific region with a high guanine-cytosine (GC) content within PHO1.2 mRNA that inhibits mRNA translation and is rearranged upon interaction with cis-NATpho1.2. Together, our results showed that dsRNA formation between OsPHO1.2 and cis-NATpho1.2 is constrained by intra-molecular secondary RNA structures. This might explain why this, and potentially other, sense-antisense pairs might evade silencing and be involved in various regulatory processes, such as enhancement of translation.
Next steps: Now, we are curious to know whether our findings are representative of other sense-antisense RNA interactions. It will be critical to study the secondary structure of other biologically relevant sense-antisense pairs, and to explore how this structure might modulate their function upon co-expression.
Rodrigo S. Reis, Jules Deforges, Romy R. Schmidt, Jos H.M. Schippers & Yves Poirier. (2021). An antisense noncoding RNA enhances translation via localised structural rearrangements of its cognate mRNA. The Plant Cell. https://academic.oup.com/plcell/advance-article/doi/10.1093/plcell/koab010/6119331