Integrated multi-omics framework of the plant response to jasmonic acid (Nature Plants)
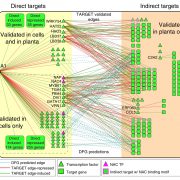
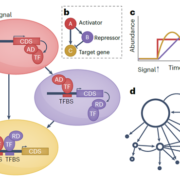
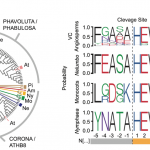
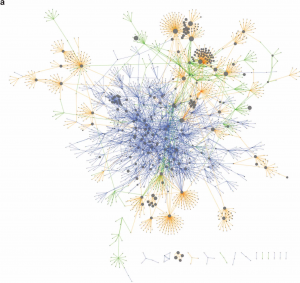 Jasmonic acid (JA) is involved in fertility, seed emergence and defense. JA perception and signal transduction are well understood, but there is limited understanding of the JA responsive genome regulatory program since only one or a small number of components are usually studied at once. Zander et al. report a multi-omics analysis of the JA regulatory network including the activities of the JA master transcription factor, MYC2, and its closest homologues. Genome-wide binding sites of MYC2 and MYC3 were identified through ChIP-seq analysis. Key findings include that almost all genes targeted by MYC3 were bound by MYC2, corroborating previous report that MYC2 and MYC3 function redundantly. Shared target genes were enriched for JA- and other hormone-related gene ontology and for DNA binding and transcriptional regulatory domains. Furthermore, these two transcription factors were also shown to directly target several hundreds of other transcription factors leading to a large gene regulatory network conferring both amplification and crosstalk. Genome-wide profiling through ChIP-seq analysis revealed MYC2’s contribution to JA-induced changes in chromatin architecture, and MYC2’s influence on the proteome and phosphoproteome was also demonstrated. Finally, a network model that predicted new components of JA signaling was generated and validated by genetic analysis. (Summary by Tolu Olukayode) Nature Plants 10.1038/s41477-020-0605-7
Jasmonic acid (JA) is involved in fertility, seed emergence and defense. JA perception and signal transduction are well understood, but there is limited understanding of the JA responsive genome regulatory program since only one or a small number of components are usually studied at once. Zander et al. report a multi-omics analysis of the JA regulatory network including the activities of the JA master transcription factor, MYC2, and its closest homologues. Genome-wide binding sites of MYC2 and MYC3 were identified through ChIP-seq analysis. Key findings include that almost all genes targeted by MYC3 were bound by MYC2, corroborating previous report that MYC2 and MYC3 function redundantly. Shared target genes were enriched for JA- and other hormone-related gene ontology and for DNA binding and transcriptional regulatory domains. Furthermore, these two transcription factors were also shown to directly target several hundreds of other transcription factors leading to a large gene regulatory network conferring both amplification and crosstalk. Genome-wide profiling through ChIP-seq analysis revealed MYC2’s contribution to JA-induced changes in chromatin architecture, and MYC2’s influence on the proteome and phosphoproteome was also demonstrated. Finally, a network model that predicted new components of JA signaling was generated and validated by genetic analysis. (Summary by Tolu Olukayode) Nature Plants 10.1038/s41477-020-0605-7
[altmetric doi=”10.1038/s41477-020-0605-7″ details=”right” float=”right”]