How to make an extraordinary machine: SMALL ORGAN 4 regulates ribosome biogenesis in plants
Ribosomes are essential molecular machines in the cell that translate mRNA sequences into proteins. Growing parts of an organism produce many ribosomes, so that after each cell division both daughter cells have enough to translate the proteins necessary for growth and development. Defects in ribosomes or in their abundance often cause lethality or diseases.
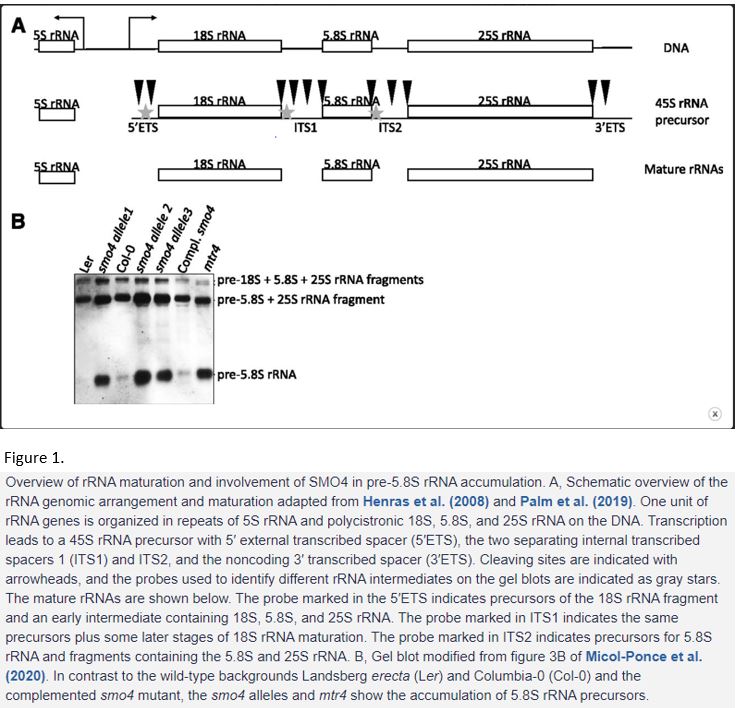 Ribosome production—or biogenesis—has been studied in various organisms. In eukaryotes, ribosomes consist of 79–80 ribosomal proteins and four main ribosomal RNAs (rRNAs), i.e., 5S, 5.8S, 18S and 25/28S, organised in two subunits (Wilson and Cate, 2012). More than 200 assembly factors and more than 70 small nucleolar RNAs are required to build a ribosome, so biogenesis is a costly process that needs to be highly regulated and quality controlled. The central part of the ribosome is built from rRNAs. In the genome, the 5S rDNA precursor is located next to a 45S precursor that contains 18S, 5.8S and 25S rDNA as a polycistronic sequence and the whole area is repeated hundreds of times (Woolford and Baserga, 2013) (Figure 1A).
Ribosome production—or biogenesis—has been studied in various organisms. In eukaryotes, ribosomes consist of 79–80 ribosomal proteins and four main ribosomal RNAs (rRNAs), i.e., 5S, 5.8S, 18S and 25/28S, organised in two subunits (Wilson and Cate, 2012). More than 200 assembly factors and more than 70 small nucleolar RNAs are required to build a ribosome, so biogenesis is a costly process that needs to be highly regulated and quality controlled. The central part of the ribosome is built from rRNAs. In the genome, the 5S rDNA precursor is located next to a 45S precursor that contains 18S, 5.8S and 25S rDNA as a polycistronic sequence and the whole area is repeated hundreds of times (Woolford and Baserga, 2013) (Figure 1A).
The 5S rDNA is transcribed by RNA polymerase III. The 45S rDNA is transcribed by RNA polymerase I and requires many steps of processing before the separate rRNAs can be integrated into pre-40S and pre-60S ribosomal particles and exported to the cytoplasm. Processing—or maturation—is a multi-step process and involves cleaving and removal of the 5′ external transcribed spacer (5’ETS), the two separating internal transcribed spacers 1 and 2 (ITS1 and 2) and the non-coding 3′ transcribed spacer (3’ETS) (Figure 1A) (Henras et al., 2008; Woolford and Baserga, 2013). There are different ways of rRNA maturation depending on which site in the spacers is cut first. Recently, researchers revealed that in plants the 5’ETS (like in yeast), ITS1 (like in human) or, in proliferating cells, ITS2 is cleaved first. A variety of plant-specific ribosome maturation factors have been discovered (Palm et al., 2019; Sáez-Vásquez and Delseny, 2019).
In this issue of Plant Physiology, Micol-Ponce and colleagues describe how the conserved protein SMALL ORGAN 4 (SMO4) acts and interacts in ribosome biogenesis (Micol-Ponce et al., 2020). SMO4 has orthologues in yeast—Nucleolar protein 53 (Nop53) and human glioma tumor-suppressor candidate region gene2 (GLTSCR2)—and regulates the maturation of rRNA. Previous studies revealed that plant SMO4 expression did not complement Nop53-deficient yeast (Zhang et al., 2015), indicating a slightly different or additional function to its orthologue despite the sequence conservation. Learning about SMO4 function will not only help us understand plant ribosome maturation better, but also potentially elucidate functions of Nop53 and GLTSCR2.
In this study, SMO4 was identified in a yeast-two-hybrid screen as an interactor of MORPHOLOGY OF ARGONAUTE1-52 SUPPRESSED2 (MAS2), a regulator of rDNA transcription (Micol-Ponce et al., 2018). Because of this interaction with a regulator of rRNA abundance and its similarity to Nop53 and GLTSCR2—both known to be involved in rRNA maturation—the authors decided that SMO4 might be involved in regulating rRNA abundance or maturation in plants as well. A further hint was the presence of a very conserved domain in SMO4 that enables interaction of Nop53 with Mtr4 in yeast. Mtr4, which has an orthologue in plants, is an RNA helicase that recruits the exosome, which degrades spacers from the 3′ end during rRNA maturation (Lange et al., 2011).
The name SMALL ORGAN 4 originates from the first mutant identified that had smaller leaves, flowers and shorter roots caused by a delay in cell division and consequently fewer but bigger cells (Zhang et al., 2015). Micol-Ponce and colleagues identified and studied several additional alleles of SMO4 and crossed the mutants to different known regulators of rRNA accumulation and maturation to assess where in the pathway SMO4 is acting. Several genes involved in rRNA maturation were found to act either synergistically with or were epistatic to SMO4 when double mutants were analyzed for typical phenotypes of rRNA knockout mutants like smaller rosette size, reduced venation patterning, and lower number and increased size of palisade mesophyll cells. Examples of such synergistically acting genes were MAS2 and NUCLEOLIN1 (NUC1), whereas RIBOSOMAL RNA PROCESSING PROTEIN 7 (RRP7) (Micol-Ponce et al., 2018) is epistatic to SMO4. MTR4 acted synergistically with SMO4 except for in venation pattering where it was epistatic to SMO4. These results confirmed that SMO4 is involved in rRNA maturation.
The SMO4 homologues Nop53 and GLTSCR2 in yeast and human, respectively, regulate 5.8S rRNA maturation. In the mutant of the SMO4 interactor MTR4, 5.8S intermediates accumulated, which indicates a function for MTR4 in 5.8S rRNA maturation too (Lange et al., 2011). To assess the presence of 5.8S rRNA intermediates in smo4, in a similar approach MicoI-Ponce et al. used probes that bind to specific spacer regions between the rRNAs characteristic for the different 5.8S rRNA intermediates in gel blots (Figure 1A). They found that in smo4 the 5.8S rRNA precursor accumulated like in mtr4 (Figure 1B). In the double mutant, they observed a synergistic effect, indicating that the two proteins might be working together but also have separate functions. Micol-Ponce and colleagues also found that nucleoli and the nucleoplasm were enlarged in smo4, the nucleolus appeared disorganised and that in ribosome profiles the portion of 40S small subunit ribosomal particles was higher. Collectively, these data indicate that SMO4 is indeed involved in rRNA maturation and its absence leads to defects in the pathway and ribosomes.
The authors conclude that some of the SMO4–MTR4 interaction previously found in yeast is conserved in plants and enables recruitment of the exosome to the 45S rRNA for proper maturation. They conclude that MTR4 and SMO4 do act together in 5.8S rRNA maturation but in different steps and that, interestingly, the rRNA maturation is not abolished when either of the factors or both are missing but is rather delayed. This delay indicates flexibility in the rRNA maturation pathway that could help keep growth and development going when some factors are not readily available.
One way to make the process more resistant to disturbance is the option to use different maturation pathways. Indeed, it has been shown recently that plants can follow different pathways for rRNA maturation (Palm et al., 2019). This represents a general principle: functional redundancy of pathways is important to enable essential processes to robustly continue in the face of disruption (Láruson et al., 2020)
While the general mechanism of ribosome biogenesis and the sequences of orthologues seem to be conserved between the kingdoms, many of the ribosome biogenesis factors have developed different functions in humans, yeast or plants. Micol-Ponce and colleagues point out that rRNA precursors do over accumulate in mutants of some Arabidopsis thaliana orthologues of yeast ribosome biogenesis factors, indicating the involvement of the proteins in rRNA maturation. On the other hand, they must be functionally different from yeast because a large subset will not complement yeast strains deficient in their respective orthologue. This is for example true for both proteins Micol-Ponce and colleagues focussed on—SMO4 and MTR4. Many of the Arabidopsis orthologues of the yeast or human ribosome maturation factors have been identified, but only a few have been analysed in detail. It will be very exciting to see more potential ribosome biogenesis factors being analysed in a systematic way with respect to their conservation and divergence. This will also help elucidate the conservation and divergence, as well as the robustness and regulation, of the whole ribosome biogenesis pathway.
Literature Cited
Henras AK, Soudet J, Gérus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y (2008) The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci 65: 2334–2359
Lange H, Sement FM, Gagliardi D (2011) MTR4, a putative RNA helicase and exosome co-factor, is required for proper rRNA biogenesis and development in Arabidopsis thaliana. Plant J 68: 51–63
Láruson ÁJ, Yeaman S, Lotterhos KE (2020) The Importance of Genetic Redundancy in Evolution. Trends Ecol Evol 35: 809–822
Micol-Ponce R, Sarmiento-Manus R, Fontcuberta-Cervera S, Cabezas-Fuster A, de Bures A, Saez-Vasquez J, Ponce MR (2020) SMALL ORGAN 4 is a ribosome biogenesis factor involved in 5.8S rRNA maturation. Plant Physiol https://doi.org/10.1104/pp.19.01540
Micol-Ponce R, Sarmiento-Mañús R, Ruiz-Bayón A, Montacié C, Sáez-Vasquez J, Ponce MR (2018) Arabidopsis RIBOSOMAL RNA PROCESSING7 is required for 18s rRNA maturation. Plant Cell 30: 2855–2872
Palm D, Streit D, Shanmugam T, Weis BL, Ruprecht M, Simm S, Schleiff E (2019) Plant-specific ribosome biogenesis factors in Arabidopsis thaliana with essential function in rRNA processing. Nucleic Acids Res 47: 1880–1895
Sáez-Vásquez J, Delseny M (2019) Ribosome biogenesis in plants: From functional 45S ribosomal DNA organization to ribosome assembly factors. Plant Cell 31: 1945–1967
Wilson DN, Cate JHD (2012) The structure and function of the eukaryotic ribosome. Cold Spring Harb Perspect Biol 4: 5
Woolford JL, Baserga SJ (2013) Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195: 643–681
Zhang X-R, Qin Z, Zhang X, Hu Y, Wenkel S (2015) Arabidopsis SMALL ORGAN 4, a homolog of yeast NOP53, regulates cell proliferation rate during organ growth. J Integr Plant Biol 57: 810–818