Insights Into How Genes are Turned On and Off in Plants
Carter et al. explore aspects of chromatin remodeling related to gene expression in Arabidopsis. https://doi.org/10.1105/tpc.17.00867
By Benjamin Carter and Joe Ogas
Background: Plant tissues are composed of cells that all contain the same genomic information and yet have very different properties. This occurs because cells selectively turn genes on or off during growth and in response to the environment. Which genes are active and which are silent depends on chemical modifications in the genome called epigenetic marks. These marks act as flags that instruct the cell’s machinery to prevent or promote transcription of a gene into RNA. For example, transcription of genes needed to produce the oily storage compounds found in seeds is promoted by activating epigenetic marks in seed cells but prevented by repressive epigenetic marks elsewhere.
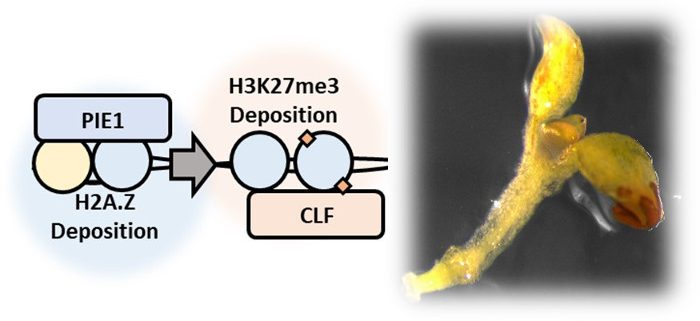
Question: We investigated whether three nuclear proteins and the epigenetic marks they promote act together or separately to control how genes are expressed in plants. These marks, called H2A.Z and H3K27me3, were previously thought to have independent roles: H2A.Z enables genes to be rapidly turned on or off whereas H3K27me3 turns genes off on a tissue-by-tissue basis.
Findings: We examined gene expression and epigenetic marks in the model plant Arabidopsis thaliana. We found that mutations in PKL, PIE1, or CLF, three nuclear proteins known to promote either H2A.Z or H3K27me3, altered expression of a common set of genes. This observation suggests that these factors act in a common gene expression pathway. Consistent with this idea, plants lacking both PIE1 and PKL show severe developmental defects. We examined where H2A.Z and H3K27me3 are located across the genome and found that 90% of H3K27me3 is located in the same places as H2A.Z. Further, we found that disabling the machinery that deposits H2A.Z (by mutating PIE1) also results in reduced levels of H3K27me3. Together, these findings indicate that the H2A.Z and H3K27me3 act in a common epigenetic pathway to control expression of genes in A. thaliana.
Next Steps: The next step is to identify the mechanisms that link these marks together in the cell. Since 90% of H3K27me3 is present in the same genomic locations as H2A.Z, we are investigating whether the presence of H2A.Z enables H3K27me3 to be deposited in the genome.
Benjamin Carter, Brett Bishop, Kwok Ki Ho, Ru Huang, Wei Jia, Heng Zhang, Pete E Pascuzzi, Roger Deal, Joe Ogas (2018). The Chromatin Remodelers PKL and PIE1 Act in an Epigenetic Pathway that Determines H3K27me3 Homeostasis in Arabidopsis. Published May 2018. DOI: https://doi.org/10.1105/tpc.17.00867
Keywords: Gene Expression, Epigenetics, Cell Identity