From Fuzziness to Clarity: Regulation of DRP5B Ring Dynamics at the Chloroplast Division Site
Tianhu Sun
ORCID ID: 0000-0002-2513-1387
Plant Breeding and Genetics Section, School of Integrative Plant Science, Cornell University, Ithaca, New York 14853
Plant cells harbor a varying number of chloroplasts and chloroplasts multiply by division to maintain the continuity of chloroplasts. Chloroplast division is performed by the constriction of two self-assembling GTPase rings in the middle of the chloroplast: the FtsZ ring on the stromal side and the DYNAMIN-RELATED PROTEIN 5B (DRP5B) ring on the cytosolic side. Chloroplasts are surrounded by inner and the outer envelope membranes (IEM and OEM) and chloroplast division is coordinated across the double membrane by the IEM-spanning protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS6 (ARC6) and its paralog PARC6 and the OEM-spanning proteins PLASTID DIVISION1 and 2 (PDV1 and PDV2) (Osteryoung and Pyke, 2014). In recent years, the identification of many key components contributing to chloroplast division in Arabidopsis thaliana (Arabidopsis) and advances in imaging technologies have helped elucidate how the division site is positioned and coordinated by the membrane division complexes (Chen et al., 2018). However, our understanding of the dynamic regulation remains fuzzy.
 DRP5B, a cytosolic dynamin protein with GTPase activity, has been long presumed to function as a molecular motor during chloroplast division (Gao et al, 2003). It is thought to provide the constriction force needed for chloroplast division by assembling a ring structure around the outside of chloroplast that finally results in chloroplast fission. In this issue of Plant Physiology, Sun et al. (2020) demonstrate that the assembly and remodeling of DRP5B ring are differentially regulated by the chloroplast OEM proteins PDV1 and PDV2.
DRP5B, a cytosolic dynamin protein with GTPase activity, has been long presumed to function as a molecular motor during chloroplast division (Gao et al, 2003). It is thought to provide the constriction force needed for chloroplast division by assembling a ring structure around the outside of chloroplast that finally results in chloroplast fission. In this issue of Plant Physiology, Sun et al. (2020) demonstrate that the assembly and remodeling of DRP5B ring are differentially regulated by the chloroplast OEM proteins PDV1 and PDV2.
Observing the dynamic regulation of DRP5B ring assembly during chloroplast division presents a number of challenges. First of all, chloroplasts are constantly mobile and frequently re-position in response to light (Kong et al., 2013), which makes live observation difficult. A mutant of the UVA/blue light photoreceptor (phot2-1) was used in this study to reduce chloroplast movement under laser scanning microscopy. The authors transformed the drp5B-1 mutant with a green fluorescent protein (GFP)-DRP5B fusion construct. This transformation restored normal chloroplast numbers and the DRP5B ring was observed on the chloroplast surface at the division site in drp5B-1/GFP-DRP5B plants. These genetic complementation experiments indicated that the GFP-DRP5B fusion protein was functional. To study subunit exchange of the assembled DRP5B ring, Sun et al. (2020) used Fluorescence Recovery After Photobleaching (FRAP), monitoring the recovery of the fluorescent signal within the bleached area. They found that the GFP-DRP5B signal recovered rapidly, indicating that the DRP5B ring undergoes active subunit exchange between the cytosolic DRP5B protein pool and the chloroplast division site.
Since DRP5B exhibits GTP hydrolysis activity, Sun et al. also asked whether this activity affects DRP5B association with the chloroplast division site or subunit exchange within the site. Two mutations in key sites of DRP5B GTPase domain, DRP5B(K61A) and DRP5B(T82D), were expressed as GFP fusions in the drp5B-1 mutant background; these proteins showed normal localization within the division site. Interestingly, when performing FRAP analysis of GFP-DRP5B and GFP-DRP5B(T82D) in the drp5B-2 phot2-1 background, a slower recovery of GFP-DRP5B(T82D) was observed. All these results indicated that GTP hydrolysis activity is not necessary for DRP5B association and assembly at the chloroplast division site but might affect subunit exchange.
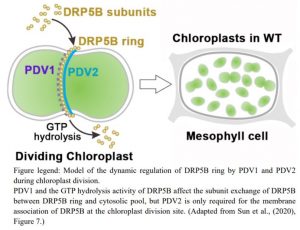
What factors could affect DRP5B association and dynamics at the chloroplast division site? PDV1 and PDV2 are the only OEM proteins known to function in chloroplast division to date (Miyagishima et al., 2006). Therefore, Sun et al. examined the localization of GFP-DRP5B in pdv1 and pdv2 mutants. Surprisingly, they found that GFP-DRP5B still associated with the chloroplast division ring in pdv1-1 mutant. However, this was not the case in pdv2 mutants, indicating that PDV2 is essential for DRP5B recruitment at the chloroplast division site. Next the GFP-DRP5B construct was transformed to pdv1-1 phot2-1 and PDV2-1/pdv2-1 phot2-1, a pdv2 heterozygote in the phot2 background, to allow DRP5B ring formation for FRAP analysis. These studies demonstrated that subunit exchange within the DRP5B ring was slowed by the loss of the PDV1 protein. However, the reduction of endogenous PDV2 protein did not affect the recovery of the DRP5B ring by subunit exchange from the cytosolic pool.
Overall, Sun et al. provide new insight into the dynamic regulation of DRP5B ring association and its assembly at the chloroplast division site. PDV1 and PDV2 were previously considered to be functionally redundant for DRP5B localization, but the authors discovered clear differences in the roles of PDV1 and PDV2 in DRP5B subunit exchange and ring assembly, respectively. This exciting study also raises a number of questions: Are there any other factors involved in DRP5B ring dynamics? How does the plant coordinate cell mitosis and chloroplast division through the regulation of DRP5B? Could chloroplast physiological state affect the DRP5B ring constriction and by what means? Clearly there is much to learn about the dynamics of chloroplast division.
LITERATURE CITED
Chen C, MacCready JS, Ducat DC, Osteryoung KW (2018). The molecular machinery of chloroplast division. Plant Physiol 176: 138-151
Gao HB, Kadirjan-Kalbach D, Froehlich JE, Osteryoung KW (2003) ARC5, a cytosolic dynamin-like protein from plants, is part of the chloroplast division machinery. Proc Natl Acad Sci USA 100: 4328-4333
Kong SG, Arai Y, Suetsugu N, Yanagida T, Wada M (2013) Rapid severing and motility of chloroplast-actin filaments are required for the chloroplast avoidance response in Arabidopsis. Plant Cell 25: 572-590
Miyagishima S, Froehlich JE, Osteryoung KW (2006) PDV1 and PDV2 mediate recruitment of the dynamin-related protein ARC5 to the plastid division site. Plant Cell 18: 2517-2530
Osteryoung KW, Pyke KA (2014) Division and dynamic morphology of plastids. Annu Rev Plant Biol 65: 443–472
Sun B, Zhang Q, Yuan H, Gao W, Han B, and Zhang M (2020). PDV1 and PDV2 differentially affect remodeling and assembly of the chloroplast DRP5B ring. Plant Physiol https://doi.org/10.1104/pp.19.01490