Finding candidate genes in maize with Camoco (Plant Cell)
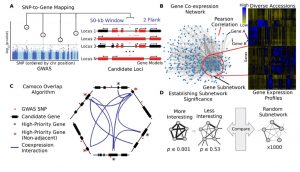 Schaefer and colleagues have developed Camoco (Co-analysis of molecular components), which integrates data from GWAS and co-expression networks to identify high confidence candidate genes associated with a phenotype of interest. To evaluate the program they used GWAS and co-expression data from maize. They used gene expression data from three datasets: data from whole-seedling transcriptome from diverse maize inbred lines, data from different maize tissues and data points from maize reference accession B73, and a dataset from roots from maize inbreds with different primary elemental absorption phenotypes. In the three cases the gene expression data came from RNA-seq, but microarray data could also be used. The evaluation showed that expression profiles from genetically diverse individuals produced more powerful co-expression networks compared to data from a single reference genotype. Two genes identify by Comoco in the root dataset were validated using mutants. The authors suggested that the identification of the best co-expression context (i.e. tissue) for a determined GWAS improves the odds of finding the genes associated to the phenotype of interest. (Summary by Cecilia Vasquez-Robinet) Plant Cell 10.1105/tpc.18.00299
Schaefer and colleagues have developed Camoco (Co-analysis of molecular components), which integrates data from GWAS and co-expression networks to identify high confidence candidate genes associated with a phenotype of interest. To evaluate the program they used GWAS and co-expression data from maize. They used gene expression data from three datasets: data from whole-seedling transcriptome from diverse maize inbred lines, data from different maize tissues and data points from maize reference accession B73, and a dataset from roots from maize inbreds with different primary elemental absorption phenotypes. In the three cases the gene expression data came from RNA-seq, but microarray data could also be used. The evaluation showed that expression profiles from genetically diverse individuals produced more powerful co-expression networks compared to data from a single reference genotype. Two genes identify by Comoco in the root dataset were validated using mutants. The authors suggested that the identification of the best co-expression context (i.e. tissue) for a determined GWAS improves the odds of finding the genes associated to the phenotype of interest. (Summary by Cecilia Vasquez-Robinet) Plant Cell 10.1105/tpc.18.00299