FERONIA holds the key to a better understanding of the molecular interplay in diverse biological processes
Hongqing Guo, Department of Genetics, Development and Cell Biology, Iowa State University
Background: FERONIA (FER) is a plasma membrane-localized receptor kinase that belongs to the CrRLK1L family, which consists of 17 members in Arabidopsis. FER, together with co-receptors LLGs/LRE and ligands such as RALF peptides, plays critical roles in plant growth, stress responses and reproduction. Better understanding the functions and underlying molecular mechanisms of FER will help illustrate how this receptor kinase balances plant growth and stress in response to environmental cues.
Question: We wished to understand the molecular mechanisms underlying FER-mediated biological processes, and to identify novel functions of FER.
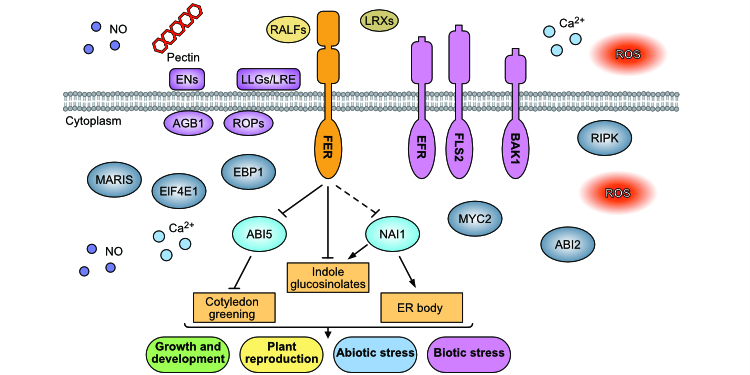 Findings: Our integrated omics study revealed that FER regulates the expression of thousands of transcripts and the abundance of thousands of proteins, as well as the phosphorylation of more than one thousand proteins. We also uncovered an extensive involvement of transcription factors in FER-mediated signaling. We identified and experimentally validated novel functions for FER in regulating endoplasmic reticulum (ER) body formation and indole glucosinolate biosynthesis. In addition, we established that FER phosphorylates and destabilizes ABI5, an important transcription factor in ABA signaling, to mediate cotyledon greening.
Findings: Our integrated omics study revealed that FER regulates the expression of thousands of transcripts and the abundance of thousands of proteins, as well as the phosphorylation of more than one thousand proteins. We also uncovered an extensive involvement of transcription factors in FER-mediated signaling. We identified and experimentally validated novel functions for FER in regulating endoplasmic reticulum (ER) body formation and indole glucosinolate biosynthesis. In addition, we established that FER phosphorylates and destabilizes ABI5, an important transcription factor in ABA signaling, to mediate cotyledon greening.
Next steps: Our multiomics analysis revealed many potential FER substrates that are involved in diverse biological processes. We will investigate the FER substrates and establish context-specific FER signaling pathways. We will also investigate how FER controls hundreds of transcription factors that regulate thousands of genes for different biological processes.
Reference:
Ping Wang, Natalie M Clark, Trevor M. Nolan, Gaoyuan Song, Parker M. Bartz, Ching-Yi Liao, Christian Montes-Serey, Ella Katz, Joanna K. Polko, Joseph J. Kieber, Daniel J. Kliebenstein, Diane C Bassham, Justin W Walley, Yanhai Yin and Hongqing Guo (2022). Integrated Omics Reveal Novel Functions and Underlying Mechanisms of FERONIA Receptor Kinase in Arabidopsis thaliana. https://doi.org/10.1093/plcell/koac111



