Father Knows Best? Small RNA Pathway Controls Endosperm Response to Paternal Genomic Dosage
In flowering plants, seed viability is dependent upon the endosperm, a triploid tissue produced by fertilization of a diploid central cell by a haploid sperm. Most endosperm genes are expressed in a 2:1 maternal to paternal ratio reflecting the genomic DNA content. Consequently, balance between maternal and paternal genome dosages is important for proper endosperm, and thus seed, development. Extra copies of the paternal genome in endosperm cells often result in seed abortion, but what determines the threshold between seed lethality and viability is unknown.
Several current models suggest different mechanisms by which endosperm ploidy incompatibilities result from epigenetic abnormalities. For example, increased expression of transposable elements (TEs) or mis-regulation of imprinted genes have been hypothesized to increase seed abortion in interploid crosses. One epigenetic mark, DNA methylation, represses expression of some endosperm genes yet promotes the expression of others (Satyaki and Gehring, 2017). Methyl marks are established and partially maintained by the RNA-directed DNA methylation (RdDM) pathway. New insight into the function of NRPD1, which encodes the largest subunit of RNA Pol IV in triploid endosperm, indicates that paternally inherited mutations in NRPD1 repress seed abortion caused by excess paternal genome dosage. A model resulting from this work hypothesizes that loss of RNA Pol IV-dependent sRNA production in the endosperm is essential for seed viability in cases of paternal genomic excess (Erdmann et al., 2017).
 To test the hypotheses generated from these models and illuminate endosperm paternal dosage effects on seed viability, Satyaki and Gehring (2019) report a comprehensive series of genetic and genomic experiments using unbalanced ploidy in Arabidopsis. To investigate the contribution of paternal RdDM pathway activity to seed viability, the authors first performed crosses between wild type diploid mothers and tetraploid fathers that were either wild type or harbored one or more mutations in RdDM pathway genes. The results suggest that paternal loss of RdDM pathway components is sufficient to suppress seed abortion caused by excess paternal genomic dosage.
To test the hypotheses generated from these models and illuminate endosperm paternal dosage effects on seed viability, Satyaki and Gehring (2019) report a comprehensive series of genetic and genomic experiments using unbalanced ploidy in Arabidopsis. To investigate the contribution of paternal RdDM pathway activity to seed viability, the authors first performed crosses between wild type diploid mothers and tetraploid fathers that were either wild type or harbored one or more mutations in RdDM pathway genes. The results suggest that paternal loss of RdDM pathway components is sufficient to suppress seed abortion caused by excess paternal genomic dosage.
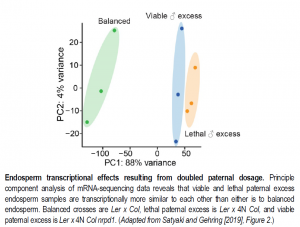
Subsequently, the authors compared global gene and TE expression, DNA methylation, and sRNA production from endosperm of viable seeds (diploid x diploid), inviable seeds (diploid mother x tetraploid father), and viable paternal excess seeds (diploid mother x tetraploid nrpd1 father). Taken together, the results indicate that paternal genomic excess induces regulatory changes at both maternal and paternal alleles and produces major changes in endosperm gene and TE expression profiles, though these changes are similar between viable and inviable seeds (see Figure). Further, loss of paternal RdDM function is associated with only modest gene expression changes. Overall, these results together with accompanying data do not support models in which endosperm ploidy incompatibilities result from mis-regulation of TEs or imprinted genes.
In addition to major changes in gene expression profiles of both viable and inviable seeds, signatures of transcriptional buffering are evident in allele-specific transcriptome data. Specifically, increases in expression of maternal alleles and repression of paternal alleles were observed in response to excess paternal genomic dosage. The outcome is that some parts of the transcriptome are more maternal than expected, thereby countering excess paternal dosage. Further work will be necessary to unravel the mechanistic basis of this buffering system and determine its relationship to the RdDM pathway.
Satyaki and Gehring (2019) prompt re-evaluation of the existing models for endosperm sensitivity to parental genome dosage. Notably, this work reveals that small expression changes instructed by RdDM-mediated paternal imprints likely determine the threshold between seed viability and abortion in cases of paternal genomic excess. Future experiments will be necessary to illuminate the specific mechanism by which seed lethality is repressed. In addition to advancing Arabidopsis research, this work provides an important foundation for understanding hybrid incompatibilities and speciation in non-model systems.
REFERENCES
Erdmann RM, Satyaki PRV, Klosinska M, and Gehring M (2017). A small RNA pathway mediates allelic dosage in endosperm. Cell Rep. 21: 3364–3372.
Satyaki PRV and Gehring M (2017). DNA methylation and imprinting in plants: machinery and mechanisms. Crit. Rev. Biochem. Mol. Biol. 52: 163–175.
Satyaki, PRV and Gehring, M (2019). Paternally-acting canonical RNA-directed DNA methylation pathway genes sensitize Arabidopsis endosperm to paternal genome dosage. Published May 2019, DOI: https://doi.org/10.1105/tpc.19.00047.