Exploring the evolution of immunity responses in Brassicaceae
By Thomas M. Winkelmüller, Frederickson D. Entila, Shajahan Anver, and Kenichi Tsuda
Background: Plants are surrounded by many microorganisms. To react to these microbes, plants have evolved receptors to sense conserved microbial molecules called microbe associated molecular patterns (MAMPs). For example, many bacteria use flagella for motility, and plants recognize the MAMP flg22 (a 22 amino-acid fragment of flagellin, a principal component of bacterial flagella). Perception of flg22 by the plant cell-surface receptor, which is conserved in most angiosperms, triggers a cascade of immune responses called pattern-triggered immunity (PTI), including transcriptional reprogramming. PTI helps plants properly respond to microbes and is well described in the model plant Arabidopsis thaliana. However, the evolution of PTI responses remains poorly explored.
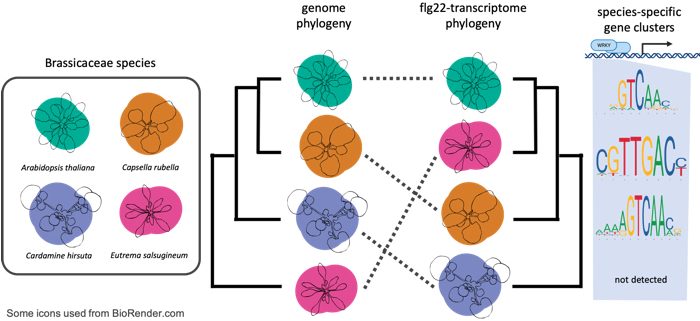
Question: How do PTI responses vary within A. thaliana accessions and across closely related Brassicaceae species? How did PTI responses such as transcriptional reprogramming evolve within a species and among Brassicaceae?
Findings: We profiled the PTI responses of selected A. thaliana accessions and three additional closely related Brassicaceae species upon flg22 treatment. Flg22 induced typical PTI responses across Brassicaceae species, although physiological consequences such as the effects of PTI on pathogen resistance and plant growth quantitatively differed. Flg22 treatment induced immediate massive transcriptional reprogramming in the Brassicaceae species, with gene clusters displaying conserved and species-specific patterns. The differences between the transcriptomic responses of A. thaliana accessions to flg22 were much smaller than those between Brassicaceae species. These differences did not reflect the phylogenic relatedness of the Brassicaceae species, suggesting that some species-specific expression patterns were under evolutionary selection. Our results also suggest that the emergence of cis-regulatory elements that are bound by WRKY transcription factors (well known to be involved in immunity) facilitated the evolution of the species-specific gene expression patterns in PTI.
Next steps: We will further investigate what drove the evolution of transcriptome responses in PTI and the evolutionary consequences. PTI plays a pivotal role in shaping local microbial communities and defending against invading pathogens. Systematic investigation of the evolution of PTI responses in the context of plant adaptation to microbes and microbial communities will advance our understanding of plant survival in diverse environments and inform agricultural practices.
Thomas M. Winkelmüller, Frederickson Entila, Shajahan Anver, Anna Piasecka, Baoxing Song, Eik Dahms, Hitoshi Sakakibara, Xiangchao Gan, Karolina Kułak, Aneta Sawikowska, Paweł Krajewski, Miltos Tsiantis, Ruben Garrido-Oter, Kenji Fukushima, Paul Schulze-Lefert, Stefan Laurent, Paweł Bednarek, and Kenichi Tsuda. (2021). Gene expression evolution in pattern-triggered immunity within Arabidopsis thaliana and across Brassicaceae species. https://doi.org/10.1093/plcell/koab073