Exosome-Deficient Mutants Reveal Rare Promoter Upstream Transcripts (PROMPTs) in Arabidopsis
In eukaryotes and archaea, protein-coding DNA is transcribed to messenger RNA (mRNA) via a preinitiation complex (PIC) composed of over 100 proteins, among them regulatory proteins, numerous transcription factors, and RNA polymerase II—the enzyme that produces precursor messenger RNA (pre-mRNA). Although transcription is traditionally represented by the synthesis of a single unidirectional transcript along the forward strand of DNA by RNA polymerase, in many eukaryotes two PICs initiate transcription divergently, causing genes to be transcribed bidirectionally from two distinct transcription start sites (TSSs) on opposite strands. Bidirectional transcription produces both a pre-mRNA transcript from the forward strand and either a second pre-mRNA transcript or one or more non-coding RNAs from the reverse strand (Wei et al., 2011).
Promoter upstream transcripts (PROMPTs) are a class of non-coding RNAs of varying lengths produced by bidirectional transcription along the reverse strand, first identified in humans (Preker et al, 2008, reviewed in Andersson and Sandelin, 2020). Like many noncoding RNA species, most PROMPTs are quickly degraded by the nuclear exosome complex, although they can also regulate gene expression. One notable feature of PROMPTs is that they appear to be considerably more common in animals than in plants, possibly due to differences in the transcription process itself.
 Here, Thieffry and colleagues (2020) used Arabidopsis (Arabidopsis thaliana) exosome-deficient mutants to show that PROMPTs are present in that system, although they are much rarer than in animals. The authors used a targeted deep RNA sequencing (RNA-seq) procedure known as Cap Analysis of Gene Expression (CAGE), in which the 5’ ends of mRNAs are preferentially sequenced. This yields a broad overview of the mRNAs present and permits the identification of TSSs with nucleotide precision. The authors first made a highly detailed map of TSSs found in wild-type plants and verified that these sites were located close to gene models present in the TAIR10 or ARAPORT11 genome annotations. Next, they obtained CAGE and RNA-seq datasets for the mutants hen2-4 (HUA enhancer 2) and rrp4-2 (ribosomal RNA processing 4), both defective in nuclear exosome function, compared them to their wild type TSS map, and analyzed the resulting forward and reverse CAGE signals. If PROMPTs are common in Arabidopsis, the forward and reverse CAGE signals would be similar in mutant plants, but not in the wild type. However, this is not what the authors observed: in general, the CAGE signal from the forward strand was considerably stronger regardless of the genetic background, suggesting that unidirectional transcription is the rule.
Here, Thieffry and colleagues (2020) used Arabidopsis (Arabidopsis thaliana) exosome-deficient mutants to show that PROMPTs are present in that system, although they are much rarer than in animals. The authors used a targeted deep RNA sequencing (RNA-seq) procedure known as Cap Analysis of Gene Expression (CAGE), in which the 5’ ends of mRNAs are preferentially sequenced. This yields a broad overview of the mRNAs present and permits the identification of TSSs with nucleotide precision. The authors first made a highly detailed map of TSSs found in wild-type plants and verified that these sites were located close to gene models present in the TAIR10 or ARAPORT11 genome annotations. Next, they obtained CAGE and RNA-seq datasets for the mutants hen2-4 (HUA enhancer 2) and rrp4-2 (ribosomal RNA processing 4), both defective in nuclear exosome function, compared them to their wild type TSS map, and analyzed the resulting forward and reverse CAGE signals. If PROMPTs are common in Arabidopsis, the forward and reverse CAGE signals would be similar in mutant plants, but not in the wild type. However, this is not what the authors observed: in general, the CAGE signal from the forward strand was considerably stronger regardless of the genetic background, suggesting that unidirectional transcription is the rule.
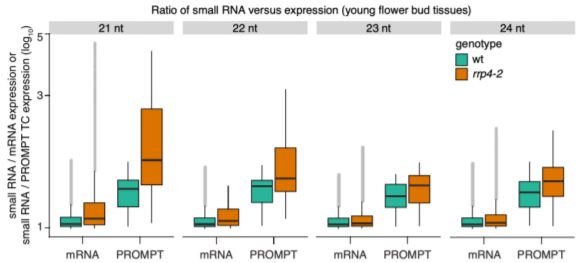
The authors also identified 96 specific TSSs for which exosome-sensitive transcripts were produced on the forward strand—i.e. PROMPTs. These TSSs were not overrepresented in any functional category, contained significant enrichment of predicted 3’ splice sites, and were distinct from previously described upstream non-coding transcripts (UNTs). Further examination of small interfering RNAs (siRNAs) produced by wild-type and exosome-deficient mutant plants showed that the Arabidopsis PROMPT regions of the mutant plants accumulated more siRNAs than the associated mRNA coding region (see Figure).
Taken together, the authors show that, although very rare, PROMPTs are detectable at individual Arabidopsis genes. Further, if allowed to accumulate, these PROMPTs are converted into siRNAs. These findings agree with recent research suggesting that PROMPTs may play a role in gene regulation despite their rarity (Song et al., 2019). However, it remains unclear whether transcription initiation on the reverse strand upstream of a TSS is uncommon in Arabidopsis, or that efficient, non-exosomic RNA degradation systems mask the production of noncoding RNAs. Future work should focus on determining how prevalent PROMPTs are in plant systems, as well as whether differences in RNA metabolism between plants and animals are responsible for differences in the prevalence of PROMPTs, or whether plants have evolved specific mechanisms of avoiding RNA interference from PROMPTs.
William Hughes
Department of Ecology, Environment, and Plant Sciences,
Stockholm University
Svante Arrhenius väg 20
114 18 Stockholm
ORCID: 0000-0003-4142-5279
REFERENCES
Andersson, R., and Sandelin, A. (2020) Determinants of enhancer and promoter activity of regulatory elements. Nat Rev Genet. 21: 71-87
Preker, P., Nielsen, J., Kammler, S., Lykke-Andersen, S., Christiansen, M., Mapendano, C., Schierup, M., and Jensen, T. (2008). RNA exosome deletion reveals transcription upstream of active human promoters. Science. 322: 1851-1854
Song, Y., Xuan, A., Bu, C., Ci, D., Tian, M., and Zhang, D. (2019). Osmotic stress-responsive promoter upstream transcripts (PROMPTs) act as carriers of MYB transcription factors to induce the expression of target genes in Populus simonii. Plant Biotech J. 17: 164-177
Thieffry, A., Vigh, M.L., Bornholdt, J., Ivanov, M., Brodersen, P. and Sandelin, A. (2020). Characterization of Arabidopsis thaliana promoter Bidirectionality and Antisense RNAs by Depletion of Nuclear RNA Decay Pathways. The Plant Cell. Published March 2020. DOI: https://doi.org/10.1105/tpc.19.00815
Wei, W., Pelechano, V., Järvelin, A., and Steinmetz, L. (2011). Functional consequences of bidirectional promoters. Trends Genet. 27: 267-276