Enhanced discovery of plant RNA-binding proteins
Yong Zhang and Devinder Sandhu; University of California Riverside and US Salinity Laboratory (USDA-ARS), Riverside, CA, USA
Background: RNA-binding proteins (RBPs) are important for controlling the fate of cells and for regulating important processes during development. Scientists study RBPs in a systematic way to understand how they control genes. However, studying RBPs in plants is difficult because plants have complex tissues. Additionally, most plant RBPs have only been discovered by studying a small fraction of RNA molecules called polyadenylated (poly(A)) RNA, which may not give a complete picture of all the RBPs. As a result, the true number of RBPs in plants may be much higher than previously thought.
Question: How can we develop a method to capture all the RBPs in plants, including those that bind to different types of RNA, i.e. poly(A) and non-poly(A), to get a comprehensive and unbiased understanding of RBPs in plants?
Findings: We developed a robust plant phase extraction (PPE) method and identified both constitutive and tissue-specific RBPs from Arabidopsis leaf and root tissues under normal growth conditions and salinity stress. We identified several previously un-annotated RBPs, some of which are involved plant responses to salt stress. More importantly, 40% of the RBPs discovered by PPE are non-poly(A) RBPs. Interestingly, many of the new RBPs are metabolic/catabolic enzymes that do not have the classic RNA-binding domains that scientists typically associate with RBPs. We also uncovered 38 domains from these enzymes as putative RNA-binding domains and provided evidence that some catalytic domains of these enzymes can directly bind RNA.
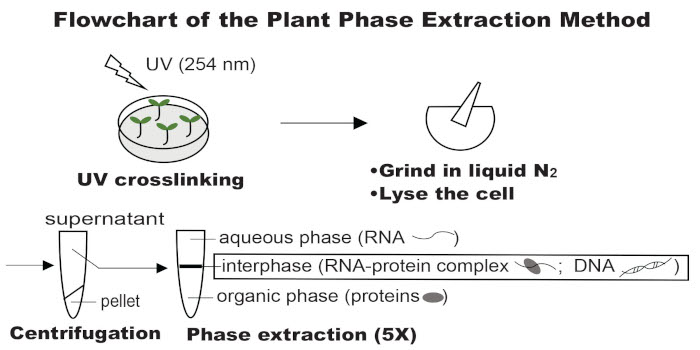
Next steps: It would be intriguing to study how these salt-responsive RBPs respond to salinity stress at the post-transcriptional level and how RNA competes or cooperates with the substrate for these metabolic/catabolic enzymes in the regulation of RNA processing and intermediary metabolism.
Reference:
Yong Zhang, Ye Xu, Todd H. Skaggs, Jorge F.S. Ferreira, Xuemei Chen and Devinder Sandhu (2023) Plant phase extraction: A method for enhanced discovery of the RNA-binding proteome and its dynamics in plants https://doi.org/10.1093/plcell/koad124