Endosidin20: A Key to Unlock the Secrets of Cellulose Biosynthesis
By now, you would think we’d know everything about cellulose, given its status as the most abundant biopolymer on Earth and its simple chemical composition. Cellulose is composed of β-1,4-D-glucose units assembled into straight chain polymers. These rod-like molecules are packed into strong microfibrils held tightly together by hydrogen bonds and van der Waals forces. Cellulose provides plant cell walls with the mechanical strength needed to resist turgor pressure while affecting the size, shape, and direction of cell growth and division, thereby shaping plant morphology (Saxena and Brown, 2005). Although both paper and cotton (Gossypium hirsutum) fibers contain over 90% cellulose, and cellulose is used to make everything from ice cream to biofuel, we still don’t understand exactly how plants synthesize this polymer. We do know that elegant cellulose synthase (CESA) complexes (CSCs) are assembled in the endoplasmic reticulum or Golgi apparatus and delivered to the plasma membrane (PM), where they actively synthesize cellulose from uridine diphosphate (UDP)-glucose in the cytosol (reviewed in Polko and Kieber, 2019). However, how CESAs catalyze cellulose synthesis is unclear, and how this catalytic activity affects CSC trafficking is unknown.
 CSCs are little machines composed of three isoforms of monomeric CESAs arranged in six-fold symmetrical rosettes at a 1:1:1 ratio. Each complex contains a cytoplasmic catalytic domain, transmembrane regions, plant-conserved sequences, and class-specific regions thought to facilitate CSC assembly and trafficking. However, the lack of tools to probe cellulose biosynthesis, such as CESA inhibitors with known modes of action, has hampered efforts to explore CSC activity. In a Breakthrough Report, Huang et al. (2020) describe the small molecule Endosidin20 (ES20), a powerful new cellulose synthesis inhibitor that directly targets the Arabidopsis thaliana (Arabidopsis) CESA isoform CESA6.
CSCs are little machines composed of three isoforms of monomeric CESAs arranged in six-fold symmetrical rosettes at a 1:1:1 ratio. Each complex contains a cytoplasmic catalytic domain, transmembrane regions, plant-conserved sequences, and class-specific regions thought to facilitate CSC assembly and trafficking. However, the lack of tools to probe cellulose biosynthesis, such as CESA inhibitors with known modes of action, has hampered efforts to explore CSC activity. In a Breakthrough Report, Huang et al. (2020) describe the small molecule Endosidin20 (ES20), a powerful new cellulose synthesis inhibitor that directly targets the Arabidopsis thaliana (Arabidopsis) CESA isoform CESA6.
A chemical library screen identified ES20, which markedly alters Arabidopsis seedling morphology by reducing cellulose levels. More than a dozen ethyl methanesulfonate-induced mutants that grew well on ES20 harbored mutations in CESA6, suggesting that ES20 directly affects CESA6 activity. Topological analysis, structural modeling, and molecular docking analysis indicated that ES20 targets the UDP-glucose binding site (i.e., catalytic site) of CESA6 to inhibit cellulose polymerization; the authors confirmed this by analyzing mutant forms of CESA6. The authors performed several biochemical assays to show that ES20 targets CESA6 directly, finding that it competes with UDP-glucose for direct binding to this CESA.
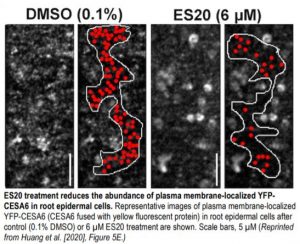
To explore whether the catalytic activity of plant CESAs affects intracellular trafficking, the authors examined the effects of ES20 on CSC localization and dynamics in transgenic seedlings expressing CESA6 fused to yellow fluorescent protein in the cesa6 loss-of-function background using high-resolution live cell imaging of root epidermal cells. ES20 treatment significantly reduced the rate of CSC motility at the PM, confirming that ES20 inhibits CSC catalytic activity. ES20 treatment also decreased the density of PM-localized CSCs (see figure), reduced the delivery of CSC to the PM, and increased the abundance of CSC at the Golgi, indicating that the catalytic activity of CESA6 affects the subcellular trafficking of CSCs. While this observation is intriguing, the relationship between CESA catalytic activity and CSC trafficking is complicated and requires further study. Perhaps ES20 could be used in conjunction with a collection of fluorescently tagged CESA6 with mutations at the catalytic site to explore the effects of CESA catalytic activity on CSC trafficking and the underlying molecular mechanisms and shed light on the mysterious process of cellulose biosynthesis.
Jennifer Lockhart
Science Editor
ORCID: 0000-0002-1394-8947
REFERENCES
Huang, L. et al. (2020). Endosidin20 targets the cellulose synthase catalytic domain to inhibit cellulose biosynthesis. Plant Cell 32: doi:10.1015/tpc00202.2020. https://doi.org/10.1105/tpc.20.00202
Polko, J.K. and Kieber, J.J. (2019). The regulation of cellulose biosynthesis in plants. Plant Cell 31, 282-296.
Saxena, I.M. and Brown, R.M. (2005). Cellulose biosynthesis: Current views and evolving concepts. Ann. Bot. 96: 9-21.



