Editor Profile: Daniel J. Kliebenstein

OLYMPUS DIGITAL CAMERA
How many genes control a phenotype? Dozens? Hundreds? Thousands? The whole genome? And what about the epigenome? Such fascinating questions on biological complexity and the interaction between genes and the environment (or, in the case of infection by a plant pathogen, multiple genomes) have long been at the heart of Daniel Kliebenstein’s research.
Studying the Whole System
Kliebenstein was born in Champaign-Urbana in east-central Illinois. As a teen he was frequently sent “on holiday” to the family farm in south-west Wisconsin, helping to lift hay bales at harvest. It was during this time that he first came to appreciate the enormous complexity of biological systems. “Driving past a field of crops it appears to be very monolithic”, he muses, “but when you actually walk through it there will be a rattle snake here and a rabbit there and you realize that it is in fact, a lot more complex.” This need to understand the whole system certainly influenced his later career path. At school, mathematics and biology were Kliebenstein’s best subjects, but he soon decided that he didn’t want to be a mathematician as he felt math was “too abstract.” Instead he studied Genetics and Chemistry at Iowa State University graduating with a BSc in 1993. It was during this time that his enthusiasm for plant biology became apparent. “I took one week of an animal science class and couldn’t even dissect a worm, so that ended that!” he laughs. And any interest in microbes was limited to studying the whole population and not to the individual organism. Hence, plants became his focus.
Following his undergraduate studies, Kliebenstein moved to Cornell University, New York to undertake his Ph.D. research in the lab of Robert Last. Here he studied the superoxide dismutase (SOD) enzymes (Kliebenstein et al., 1998) and contributed to the successful cloning of the Arabidopsis thaliana UV-B RESISTANCE LOCUS 8 (UVR8) photoreceptor (Kliebenstein et al., 2002; “a paper nobody knows I’m on!” he admits wryly). Both of these publications from his PhD research have gone on to become seminal papers in their fields. Indeed, Kliebenstein et al. (1998) has received over 500 citations and remains, from a total of over 160 peer-reviewed publications, his most highly cited paper to date. During his time as a graduate student, Kliebenstein spent a years’ sabbatical in Jeff Dangl’s plant immunology lab at the University of North Carolina. With Dangl he continued his research on Arabidopsis SODs, and more specifically, the pathogen-induced SA-dependent induction of a CuZn SOD by the negative regulator of cell death, LESION SIMULATING DISEASE 1 (Kliebenstein et al., 1999).
Model Systems: Glucosinolates and Botrytis cinerea
After completing his Ph.D. in Genetics in 1999 and identifying a need to fill in the gaps in his skills in both technical and analytical chemistry as well as statistics and quantitative biology, Kliebenstein moved across the pond and joined the newly established Max Planck Institute of Chemical Ecology in Jena, Germany. At Max Planck he was fortunate to have two mentors, the evolutionary ecologist Tom Mitchell-Olds and the biochemist Jonathan Gershenzon, an expert in plant secondary metabolites. Over the ensuing two years, Kliebenstein, Mitchell-Olds, Gershenzon and co-workers published a plethora of papers mapping quantitative trait loci (QTLs) responsible for the natural genetic variation in Arabidopsis glucosinolate profiles. Glucosinolates are a large group of important secondary metabolites that alter both insect herbivory and fungal survival and are thus vital to the plant defense response – often referred to as “the mustard oil bomb” (Ratzka et al., 2002). During this time, Kliebenstein was one of the pioneers in this field that elucidated the identity of the major polymorphic genes responsible for the multivariate glucosinolate phenotype observed in Arabidopsis. Crucially, these genes have been shown to act both additively and epistatically (Kliebenstein et al., 2001), and to be linked to fitness variation in the field, suggesting that they are adaptive (Kerwin et al.).
Kliebenstein used this time as a post-graduate researcher at Max Planck to truly make his mark in the field of quantitative genetics. However, when reflecting on this period he also acknowledges “My biggest career regret was not even submitting a couple of papers to Nature or Science about cloning QTLs for metabolites and insects in 2000. The problem is that…[you are judged on] what you are capable of based on what you publish as a graduate student and post-doc.”
His earlier sabbatical in Prof Dangl’s lab also influenced Kliebenstein’s choice of plant pathogen system going forward. He recalls, “I learnt that there were lots of people working on ETI [effector-triggered immunity], PTI [PAMP-triggered immunity], MAMPs, PAMPs, DAMPs, HAMPs… [microbial-, pathogen-, damage-, herbivore- associated molecular patterns, respectively]. I was always more interested in the biochemistry and how metabolites might act as mediators of the interaction. At the time it was already known that necrotrophs use toxins but much less was known about how one organism [e.g. Botrytis cinerea] could have individual isolates that are capable of infecting basically any land plant. The question became “How does anything apply to a pathogen that contravenes all the rules of what everyone else is studying?’” Further evidence of his fascination with all things complex!
Complex Systems: Genotype-to-Phenotype
In 2001 Kliebenstein started his own research group at the University of California, Davis where he currently resides as a full Professor and Chair of the Plant Biology Graduate Group. His research continued to utilize the innate variation in plant glucosinolate profiles as a model system to study the role of epistasis (gene x gene interactions) and environmental conditions (genotype x environment interactions) in adaptive evolution. The glucosinolate system, Kliebenstein explains “has benefits in terms of being cheap and easy; you can torture it and manipulate it and it’s not usually lethal in the lab so you can do lots of things to it. It lets me ask questions that are much bigger than the system, for example, ‘How many genes control an individual phenotype?’ Fundamental stuff like ‘What is epistasis?’, ‘What is pleiotropy?’”. Using this model system Kliebenstein’s lab has refined both QTL analysis and modern quantitative tools such as genome wide association (GWA) mapping to identify numerous novel genes and alleles responsible for fitness in the field (Chan et al., 2011).
Crucially, Kliebenstein’s group empirically demonstrated the significant contribution made by the cytoplasmic genomes of the plastids and mitochondria to phenotypic variation, despite them only containing around 1% of the number of genes found in the nuclear genome. Their own previous work on the plant metabolome had revealed extensive epistasis between nuclear loci (Rowe et al., 2008). In their seminal 2013 eLife paper, Bindu Joseph and co-workers showed that cytoplasmic genetic variation has a significant and widespread influence on the plant metabolome, but also that cyto-nuclear epistasis plays a key role in controlling this observed variation (Joseph et al., 2013). Previously, quantitative analysis had largely been restricted to the study of the nuclear genome so this paper will shape how future experiments were conducted, having as Kliebenstein himself puts it, “stealth impact”!
In addition to their model glucosinolate system, Kliebenstein’s research group is interested in the interaction between Arabidopsis and Botrytis cinerea, a necrotrophic fungus which also produces secondary metabolites that act to kill plant cells. In the mid 2000’s in association with Katherine Denby, a colleague from his time at Cornell, Kliebenstein demonstrated that Botrytis susceptibility was inversely correlated with accumulation of the major Arabidopsis phytoalexin, camalexin, but, crucially, that B. cinerea isolates contain genetic variation for sensitivity to the compound (Denby et al., 2004; Kliebenstein et al., 2005). Kliebenstein cites this 2005 publication as one of his two favorite papers (alongside the aforementioned 2013 work on the importance of the cytoplasmic genomes), he explains, “Up until then people were fighting over whether phytoalexins matter; phytoalexins don’t matter! We said that if you use a resistant pathogen they don’t matter but if you use a sensitive one they do.” Kliebenstein later went on to further identify numerous Arabidopsis QTLs contributing to variation in B. cinerea susceptibility (Rowe and Kliebenstein, 2008).
Systems Biology
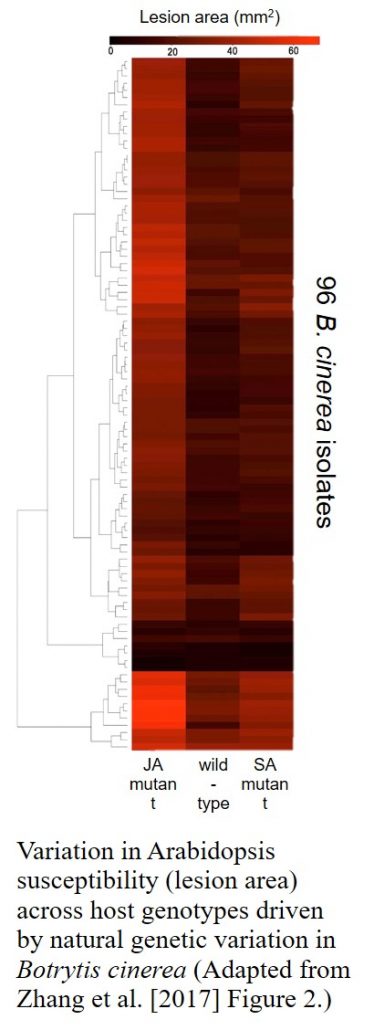 Never one to shy away from asking bold questions, Kliebenstein has more recently used the Arabidopsis-Botrytis pathosystem to show that the plant’s defense response is extremely sensitive to genetic variation in the pathogen. Arabidopsis wild-type and hormone-signaling mutants were infected with 96 genetically diverse B. cinerea isolates and the resultant host defense-related phenotypes and transcriptomic responses determined. This research and the huge underlying datasets were published late last year in The Plant Cell (Zhang et al., 2017). He is currently expanding this 96-isolate experimental system into domesticated crop species including potato, lettuce and soybean – “the nastier [more complex] I can make the experiment the happier I am”, he laughs! Indeed, such large-scale systems biology approaches and causal network models aiming to elucidate the quantitative interactions between two different genomes now form the core of his research program.
Never one to shy away from asking bold questions, Kliebenstein has more recently used the Arabidopsis-Botrytis pathosystem to show that the plant’s defense response is extremely sensitive to genetic variation in the pathogen. Arabidopsis wild-type and hormone-signaling mutants were infected with 96 genetically diverse B. cinerea isolates and the resultant host defense-related phenotypes and transcriptomic responses determined. This research and the huge underlying datasets were published late last year in The Plant Cell (Zhang et al., 2017). He is currently expanding this 96-isolate experimental system into domesticated crop species including potato, lettuce and soybean – “the nastier [more complex] I can make the experiment the happier I am”, he laughs! Indeed, such large-scale systems biology approaches and causal network models aiming to elucidate the quantitative interactions between two different genomes now form the core of his research program.
Future Systems?
To return to the question posed at the start, for this underpins much of quantitative biology, how many genes control a phenotype? Obviously, we simply don’t know! But neither can we even start to guess at the shape of that space, and it is this complexity that continues to drive Kliebenstein’s research. And in a world of increasing population and changing climate, the answers to such questions will have a huge impact on our future food security. To use a real-world example from Kliebenstein himself – imagine if two pathogens attacked a plant at the same time, in the presence of drought. What does the plant “see”? What about the pathogen? How can we tease apart these interactions to identify robust fitness phenotypes? Ultimately this comes down to the question of whether genes interact additively or epistatically – a question that has been asked for over 100 years but is yet to be resolved to a satisfactory level simply because the experimental system required to do so is too complex to fathom. Kliebenstein sums it up well when he says “Probably the most important biological questions are the ones we are afraid to ask, or don’t know we should be asking. But I think we don’t know we should be asking them because we either assume they’re known, or we can’t contemplate how to ask them”.
Early Warning Systems
Reflecting over his industrious career, what wisdom would Kliebenstein impart upon his younger self and indeed other early-career researchers? “Never forget that scientists are as messed up and illogical as everyone else. That idea that they are empirical logicians and this cold empiricism dominates any internal politics, social mores and emotion is just wrong. Scientists are way more Bones than they are Spock”.
Kliebenstein is currently based at the Department of Plant Sciences at UC Davis’ College of Agricultural and Environmental Sciences. He has served on the Editorial Board of The Plant Cell since 2011 and a Senior Editor since 2015.
Emily Breeze
School of Life Sciences, University of Warwick, Coventry, UK
References
Chan, E.K.F., Rowe, H.C., Corwin, J.A., Joseph, B., and Kliebenstein, D.J. (2011). Combining Genome-Wide Association Mapping and Transcriptional Networks to Identify Novel Genes Controlling Glucosinolates in Arabidopsis thaliana. PLoS Biol. 9: e1001125–19.
Denby, K.J., Kumar, P., and Kliebenstein, D.J. (2004). Identification of Botrytis cinerea susceptibility loci in Arabidopsis thaliana. Plant J. 38: 473–486.
Joseph, B., Corwin, J.A., Li, B., Atwell, S., and Kliebenstein, D.J. (2013). Cytoplasmic genetic variation and extensive cytonuclear interactions influence natural variation in the metabolome. eLife 2: 529–21.
Kerwin, R., Feusier, J., Corwin, J., Rubin, M., Lin, C., Muok, A., Larson, B., Li, B., Joseph, B., Francisco, M., Copeland, D., Weinig, C., Kliebenstein, D.J. (2015) Natural genetic variation in Arabidopsis thaliana defense metabolism genes modulates field fitness. eLife 4:e05604
Kliebenstein, D.J., Dietrich, R.A., Martin, A.C., Last, R.L., and Dangl, J.L. (1999). LSD1 Regulates Salicylic Acid Induction of Copper Zinc Superoxide Dismutase in Arabidopsis thaliana. Mol. Plant Microbe Interact. 12: 1022–1026.
Kliebenstein, D.J., Lambrix, V.M., Reichelt, M., Gershenzon, J., and Mitchell-Olds, T. (2001). Gene duplication in the diversification of secondary metabolism: tandem 2-oxoglutarate-dependent dioxygenases control glucosinolate biosynthesis in Arabidopsis. Plant Cell 13: 681–693.
Kliebenstein, D.J., Lim, J.E., Landry, L.G., and Last, R.L. (2002). Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant Physiol. 130: 234–243.
Kliebenstein, D.J., Monde, R.A., and Last, R.L. (1998). Superoxide dismutase in Arabidopsis: an eclectic enzyme family with disparate regulation and protein localization. Plant Physiol. 118: 637–650.
Kliebenstein, D.J., Rowe, H.C., and Denby, K.J. (2005). Secondary metabolites influence Arabidopsis/Botrytis interactions: variation in host production and pathogen sensitivity. Plant J. 44: 25–36.
Ratzka, A., Vogel, H., Kliebenstein, D.J., Mitchell-Olds, T., and Kroymann, J. (2002). Disarming the mustard oil bomb. Proc. Natl. Acad. Sci. USA 99: 11223–11228.
Rowe, H.C. and Kliebenstein, D.J. (2008). Complex Genetics Control Natural Variation in Arabidopsis thaliana Resistance to Botrytis cinerea. Genetics 180: 2237–2250.
Rowe, H.C., Hansen, B.G., Halkier, B.A., and Kliebenstein, D.J. (2008). Biochemical Networks and Epistasis Shape the Arabidopsis thaliana Metabolome. Plant Cell 20: 1199–1216.
Zhang, W., Corwin, J.A., Copeland, D., Feusier, J., Eshbaugh, R., Chen, F., Atwell, S., and Kliebenstein, D.J. (2017). Plastic Transcriptomes Stabilize Immunity to Pathogen Diversity: The Jasmonic Acid and Salicylic Acid Networks within the Arabidopsis/ BotrytisPathosystem. Plant Cell 29: 2727–2752.