Double Crossed: CDKG1 Regulates Crossover Formation by Stabilizing Meiotic and Somatic Recombination Intermediates
Genetic recombination is the process by which novel genotypes are generated from the genetic material of both parents. In eukaryotes, homologous chromosomes synapse and recombine during meiotic prophase I, during which double-stranded breaks (DSBs) are formed by DNA topoisomerases such as SPO11. If large enough, some of the recombinase-associated D-loops arising from these DSBs can form heteroduplex DNA structures known as double Holliday Junctions (dHJs). The resolution of dHJs can result in the generation of novel chromosomal configurations, termed “crossover” (CO) events, as well as the restoration of the original configuration, termed “non-crossover” (NCO) events (Blary and Jenczewski, 2019). In addition, COs can be further divided into two groups: class I COs, which are more common, depend on tightly regulated interactions with ZMM proteins, and can only occur beyond a minimum distance from other COs; and class II COs, which occur randomly and do not depend on ZMM proteins.
The selective advantage of novel genotypes arising from COs permit the combination of advantageous mutations present on different parental genomes resulting in novel genotypes, and have therefore been hypothesized to be a major benefit of sexual reproduction (Barton and Charlesworth 1998). However, the proportion of dHJs that end up forming COs (as opposed to NCOs) is relatively small (~4-7%; see Mercier et al. 2015). Which factors are involved in CO designation is obscure, despite the identification of numerous genes that resolve DSBs as NCOs.
Cyclin-dependent kinase G1 (CDKG1) was first identified as a cyclin-dependent regulator of the early G1 phase in Arabidopsis thaliana (Menges et al., 2005) and was found to be structurally related to Ph1 locus kinases responsible for homologous chromosome pairing in wheat (Triticum aestivum). Recent work has established that at high ambient temperature, CDKG1 is necessary for CO designation and normal synapsis in male meiosis (Zheng et al. 2014). The cdkg1-1 mutant, which possesses a null allele of the CDKG1 gene, showed compromised synapsis and fewer CO events at ambient temperatures above 25°C, resulting in low fertility.
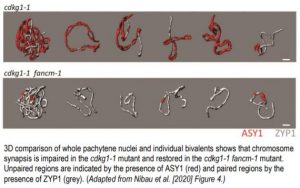 In this issue, Nibau et al. (2020) determined the precise effect of CDKG1 on recombination and synapsis by comparing the cdkg1-1 mutant to other A. thaliana mutants possessing well-characterized defects at different stages of crossover formation and junction resolution. They first crossed the cdkg1-1 mutant with the msh5-2 mutant, which contains a transfer DNA (t-DNA) insertion in AtMSH5, a ZMM protein implicated in the class I CO pathway. The cdkg1-1 msh5-2 double mutant showed an increase in bivalent formation at metaphase I, suggesting that the loss of CDKG1 results in an approximately threefold increase in the number of class II CO events. Next, the authors crossed cdkg1-1 and fancm-1 mutant plants to obtain a double mutant. The fancm-1 mutant contains a t-DNA insertion in FANCM, a helicase that negatively regulates bivalent formation via a ZMM-independent pathway (Crismani et al., 2012). Full chromosome synapsis at the pachytene substage of meiotic prophase was observed in Col-0 (Columbia) wild-type and fancm-1 plants, while—as noted in Zheng et al (2014)—synapsis fails in the cdkg1-1 mutant. However, the cdkg1-1 fancm-1 double mutant shows nearly normal synapsis, likely due to the presence of a greater number of early recombination intermediates (see Figure 1 for a 3D model of fully synapsed bivalents). However, the authors also noted that many recombination intermediates were incorrectly resolved in the double mutant, and analysis of a triple cdkg1-1 fancm-1 msh5-2 triple mutant confirmed that these intermediates were produced by class II CO pathways rather than by the class I pathway. Finally, the authors showed that CDKG1 is necessary to stabilize recombination intermediates produced during homologous recombination (HR) in somatic tissue. They did so by crossing the cdkg1-1 mutant with the reporter line IC9, which permits the detection and quantification of individual DNA repair events by somatic homologous recombination restoring a colour marker gene. The cdkg1-1 mutant showed fewer instances of spontaneous recombination events than the IC9 control line. In addition, the cdkg1-1 mutant also showed impaired HR DNA repair in response to exposure to the DSB-causing agent bleomycin.
In this issue, Nibau et al. (2020) determined the precise effect of CDKG1 on recombination and synapsis by comparing the cdkg1-1 mutant to other A. thaliana mutants possessing well-characterized defects at different stages of crossover formation and junction resolution. They first crossed the cdkg1-1 mutant with the msh5-2 mutant, which contains a transfer DNA (t-DNA) insertion in AtMSH5, a ZMM protein implicated in the class I CO pathway. The cdkg1-1 msh5-2 double mutant showed an increase in bivalent formation at metaphase I, suggesting that the loss of CDKG1 results in an approximately threefold increase in the number of class II CO events. Next, the authors crossed cdkg1-1 and fancm-1 mutant plants to obtain a double mutant. The fancm-1 mutant contains a t-DNA insertion in FANCM, a helicase that negatively regulates bivalent formation via a ZMM-independent pathway (Crismani et al., 2012). Full chromosome synapsis at the pachytene substage of meiotic prophase was observed in Col-0 (Columbia) wild-type and fancm-1 plants, while—as noted in Zheng et al (2014)—synapsis fails in the cdkg1-1 mutant. However, the cdkg1-1 fancm-1 double mutant shows nearly normal synapsis, likely due to the presence of a greater number of early recombination intermediates (see Figure 1 for a 3D model of fully synapsed bivalents). However, the authors also noted that many recombination intermediates were incorrectly resolved in the double mutant, and analysis of a triple cdkg1-1 fancm-1 msh5-2 triple mutant confirmed that these intermediates were produced by class II CO pathways rather than by the class I pathway. Finally, the authors showed that CDKG1 is necessary to stabilize recombination intermediates produced during homologous recombination (HR) in somatic tissue. They did so by crossing the cdkg1-1 mutant with the reporter line IC9, which permits the detection and quantification of individual DNA repair events by somatic homologous recombination restoring a colour marker gene. The cdkg1-1 mutant showed fewer instances of spontaneous recombination events than the IC9 control line. In addition, the cdkg1-1 mutant also showed impaired HR DNA repair in response to exposure to the DSB-causing agent bleomycin.
Taken together, Nibau et al. (2020) have established that CDKG1 is necessary for the stabilization of recombination intermediates during both male meiosis and somatic homologous recombination following DNA damage. The loss of CDKG1 results in impaired synapsis and fewer class I COs. In addition, the cdkg1-1 mutant showed three times as many class II CO events, possibly because recombination intermediates that would normally be stabilized by CDKG1 entered non-ZMM pathways instead. Finally, CDKG1 was also found—like other major genes regulating recombination such as FANCM–to play an important role in somatic DNA repair. Other studies have found that CDKG1 is involved in temperature-specific splicing cascades (Cavallari et al. 2018), stimulating speculation regarding the possible mechanism by which it may act. Thus, further work on the molecular mechanisms by which CDKG1 regulates meiotic proteins might reveal additional facets of an important protein.
William Hughes
Department of Ecology, Environment, and Plant Sciences, Stockholm University,
Svante Arrhenius väg 20, 11418 Stockholm, SE
REFERENCES
Blary, A. and Jenczewski, E. (2019) Theor. Appl. Genet. 132: 575-592.
Barton, N. and Charlesworth, B. (1998) Science 281: 1986-1990.
Cavallari, N., Nibau, C., Fuchs, A., Dadrou, D., Barta, A., and Doonan, J. (2018) Plant J. 94: 1010-1022.
Crismani, W., Girard, C., Froger, N., Pradillo, M., Santos, J., Chelsheva, L., Copenhaver, G., Horlow, C., and Mercier, R. (2012) Science 336: 1588-1590.
Doonan, J. and Kitsios, G. (2009) Mol. Biotech. 42: 14-29.
Menges, M., de Jager, S., Gruissem, W., and Murray, J. (2005) Plant J. 41: 546-566.
Mercier, R., Mezard, C., Jenczeweski, E., Macaisne, N., and Grelon, M. (2015) Ann. Rev. Plant Biol. 66: 297-327.
Nibau, C., Lloyd, A., Dadrou, D., Betekhtin, A., Tsilimigka, F., Phillips, D., and Doonan, J. (2020). CDKG1 is required for meiotic and somatic recombination intermediate processing in Arabidopsis. Plant Cell https://doi.org/10.1105/tpc.19.00942.
Zheng, T., Nibau, C., Phillips, D., Jenkins, G., Armstrong, S., and Doonan, J. (2014) Proc. Nat. Acad. Sci. USA 111: 2182-2187.




