Discovery of mitochondrial endonucleases
Prokaryotic endosymbiont-derived organelles (i.e. mitochondria and chloroplasts), contain their own genomes and the copy number of organelle genomes per cell is high; indeed, a previous study calculated that in Arabidopsis thaliana, each cell has 1000 to 1700 copies of the chloroplast genome (Zoschke et al., 2007). Moreover, the copy number changes drastically under specific conditions; for example, in plants, the copy number of organelle genomes declines during pollen development (Nagata et al., 1999). This reduction of organelle genome content in male reproductive cells during fertilization may be responsible for the maternal inheritance of the organelle genome in plants and animals (Greiner et al., 2015).
Emerging research has begun to identify the nucleases that reduce genome copy number. A previous study found an exonuclease termed DEFECTIVE IN POLLEN ORGANELLE DNA DEGRADATION1 (DPD1), which degrades the mitochondrial genome (mtDNA) and the chloroplast genome after mitosis during pollen cell development (Matsushima et al., 2011). This exonuclease removes a nucleotide from the end of a polynucleotide; therefore, endonuclease activity, which cuts the phosphodiester bond within a polynucleotide chain, is required to digest mtDNA, which is circular (Hudson and Vinograd, 1967). However, such an enzyme has not been identified in plant species. In the current issue of Plant Physiology, Ma et al. searched for a mitochondrial endonuclease through a multi-step, biochemical purification based on in-gel detection of endonuclease activity (Ma et al., 2018). As the starting material, they used isolated mitochondria from more than 10 kg of fresh maize (Zea mays) pollen. This heroic effort was successful, as they purified a 20-kD polypeptide, namely M20 (Fig. 1A). This protein has a His-Asn-His/Asn (H-N-H/N) domain that is a conserved motif of endonuclease. Both purified M20 and recombinant M20 showed endonuclease activity in vitro, confirming that M20 is indeed an endonuclease.
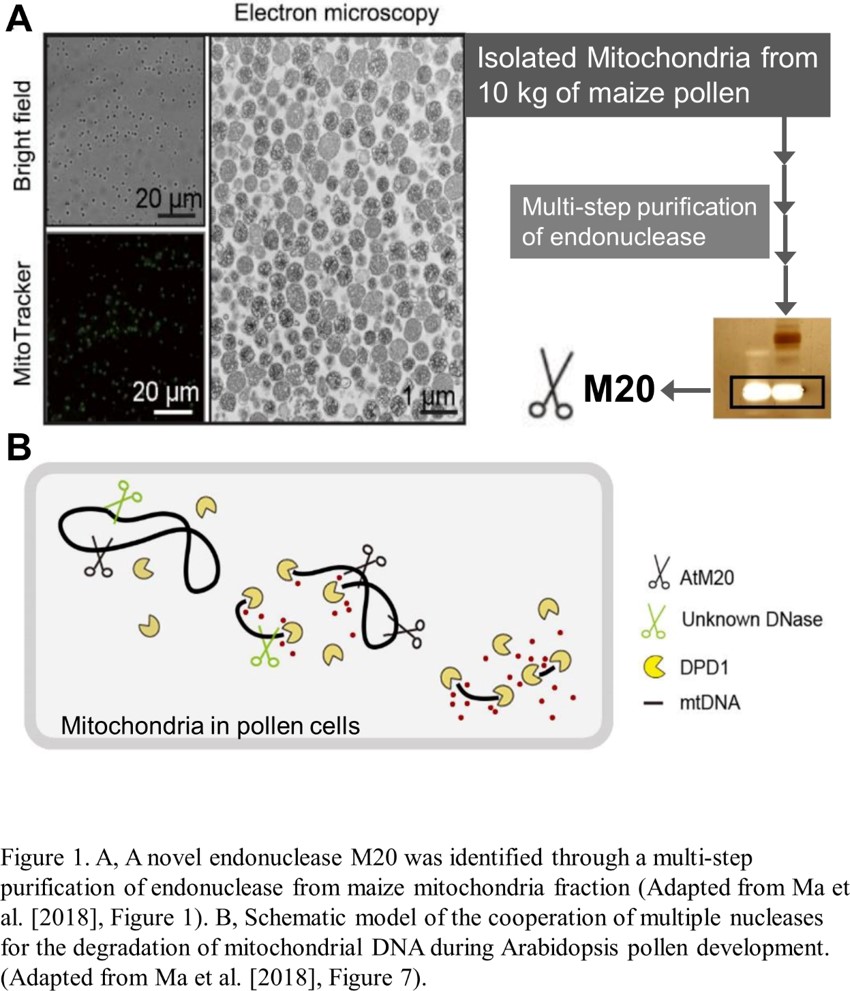
Database searches revealed that M20 is evolutionarily conserved in the plant kingdom, and the authors found the Arabidopsis thaliana M20 homolog. The Arabidopsis M20 transcript is upregulated during pollen mitosis and the M20 localizes in mitochondria. To evaluate the physiological importance of M20 in Arabidopsis, the authors generated a knockout mutant line by using CRISPR/Cas9. In the m20 mutants, the decrease in mtDNA content during pollen development was suppressed compared to wild-type plants. Overall, the study provides robust evidence indicating that M20 is an endonuclease that degrades mtDNA after pollen mitosis.
How do M20 and DPD1 cooperate during pollen development? An in vitro assessment of digestion activity for circular DNA suggested a model in which M20 endonuclease-mediated fragmentation of circular DNA is an earlier event enabling the digestion of polynucleotide fragments by exonuclease activity of DPD1 (Fig. 1B). However, in the m20 mutant plants, the mtDNA reduction during pollen development was not completely compromised. More mtDNA persisted in the dpd1 mutants than in m20 mutants. Therefore, if the schematic model of mtDNA degradation represented by Ma et al. is true in vivo (Fig. 1B), an additional, unknown endonuclease should work to support mtDNA digestion in developing pollen. A purified 30-kD polypeptide, named M30, another candidate mitochondrial endonuclease, which might act as an additional player in mtDNA breakdown in pollen. Clearly, beyond the in vitro assays, orchestration of multiple nucleases for organelle genome degradation needs to be assessed in vivo for the evaluation of mtDNA content in the double mutants of AtM20 and DPD1.
Organelle genome degradation may play an important role in various developmental processes in addition to pollen development. For example, the DPD1 transcript is upregulated in aging Arabidopsis leaves (Sakamoto and Takami, 2014). In leaf senescence, cellular components are actively degraded to facilitate the redistribution of amino acids, nucleotides, and lipids to young organs (Gan and Amasino, 1997). Since DNA is a reservoir of phosphorus, a major essential element for plant growth that is usually supplied by fertilizer application in agricultural setting, nuclease-mediated degradation of organelle genomes might act as a recycling system for phosphorus and for nucleosides and nucleobases to be reused for de novo synthesis of DNA (Zrenner et al., 2006). Does nucleotide turnover facilitate nutrient recycling during developmental aging or phosphorus starvation? This question has not been answered although the vital role of the remodeling of membrane-bound phospholipids in plant response to phosphorus starvation is known (Tjellstrom et al., 2008). The discovery of a novel mitochondrial endonuclease M20 and the establishment of its mutant line will allow researchers to address as yet uncharacterized roles of nuclease-mediated DNA digestion. The current finding is thus an important step to extend our knowledge of the mechanism for organelle genome degradation and its physiological relevance in plant growth.
LITERATURE CITED
Gan SS, Amasino RM (1997) Making sense of senescence – Molecular genetic regulation and manipulation of leaf senescence. Plant Physiol 113: 313-319
Greiner S, Sobanski J, Bock R (2015) Why are most organelle genomes transmitted maternally? Bioessays 37: 80-94
Hudson B, Vinograd J (1967) Catenated circular dna molecules in Hela cell mitochondria. Nature 216: 647-652
Matsushima R, Tang LY, Zhang L, Yamada H, Twell D, Sakamoto W (2011) A conserved, Mg2+-dependent exonuclease degrades organelle DNA during Arabidopsis pollen development. Plant Cell 23: 1608-1624
Nagata N, Saito C, Sakai A, Kuroiwa H, Kuroiwa T (1999) The selective increase or decrease of organellar DNA in generative cells just after pollen mitosis one controls cytoplasmic inheritance. Planta 209: 53-65
Sakamoto W, Takami T (2014) Nucleases in higher plants and their possible involvement in DNA degradation during leaf senescence. J Exp Bot 65: 3835-3843
Tjellstrom H, Andersson MX, Larsson KE, Sandelius AS (2008) Membrane phospholipids as a phosphate reserve: the dynamic nature of phospholipid-to-digalactosyl diacylglycerol exchange in higher plants. Plant Cell Environ 31: 1388-1398
Zoschke R, Liere K, Börner T (2007) From seedling to mature plant: Arabidopsis plastidial genome copy number, RNA accumulation and transcription are differentially regulated during leaf development. Plant J 50: 710-722
Zrenner R, Stitt M, Sonnewald U, Boldt R (2006) Pyrimidine and purine biosynthesis and degradation in plants. Annu Rev Plant Biol 57: 805-836