Designer PPR Proteins as Tools to Explore RNA Binding in vivo
Techniques to isolate the RNA molecules bound to a specific protein, via co-immunoprecipitation, for instance, have been available for years. The converse—methods to identify the proteins bound to a specific RNA—have been harder to come by, in part because of difficulty in elucidating the determinants of RNA binding specificity (reviewed in Helder et al., 2016). Now, though, our understanding of the basis for RNA binding specificity by pentatricopeptide repeat (PPR) and Pumilio/FBF (PUF) domain proteins has progressed to the point that it has been possible to change their binding specificity in a predictable fashion via directed mutations (reviewed in Hall, 2017). Furthermore, in a new Breakthrough Report, McDermott et al. (2019) demonstrate that ‘designer’ RNA binding proteins useful for isolating specific RNA sequences in vivo are within reach.
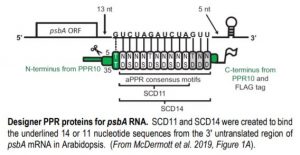
PPR proteins consist of consecutive PPR motifs, each of which binds a single nucleotide with a specificity that is strongly influenced by the identity of the amino acids at several positions within the motif (Barkan et al., 2012). Building on previous results, McDermott and coauthors explored whether an artificial PPR (aPPR) protein could be used in vivo to specifically bind psbA mRNA, a chloroplast transcript that is translationally regulated by light via unknown mechanisms. The SCD11 and SCD14 proteins contain aPPR domains that were designed to bind psbA mRNA (see Figure) and have been shown to exhibit specific binding in vitro (Miranda et al., 2018). McDermott et al. generated transgenic Arabidopsis thaliana plants expressing FLAG-tagged versions of these proteins in the chloroplasts and assessed their RNA binding in vivo. The authors isolated chloroplasts from these plants, immunoprecipitated SCD11 and SCD14 via the FLAG tag, and purified the co-immunoprecipitated RNA.
 McDermott et al. assessed the binding specificity of these proteins using RNA coimmunoprecipitation-sequencing (RIP-seq): psbA mRNA was enriched 100-fold in anti-FLAG immunoprecipitates from SCD11 and SCD14 plants versus wild type, whereas most other RNA levels showed no difference. Thus, these aPPRs bind the target RNA specifically in vivo. There was some low-level off-target binding, mostly to RNAs that shared sequence similarity with the target binding site in psbA. Importantly, the scope of this off-target binding in vivo was similar to previous in vitro results, suggesting that in vitro experiments can be informative in the design stage for aPPRs.
McDermott et al. assessed the binding specificity of these proteins using RNA coimmunoprecipitation-sequencing (RIP-seq): psbA mRNA was enriched 100-fold in anti-FLAG immunoprecipitates from SCD11 and SCD14 plants versus wild type, whereas most other RNA levels showed no difference. Thus, these aPPRs bind the target RNA specifically in vivo. There was some low-level off-target binding, mostly to RNAs that shared sequence similarity with the target binding site in psbA. Importantly, the scope of this off-target binding in vivo was similar to previous in vitro results, suggesting that in vitro experiments can be informative in the design stage for aPPRs.
Having established that these aPPRs bind psbA mRNA in vivo, the authors went on to use it to determine what endogenous proteins are bound to psbA mRNA. Mass spectrometry analysis of the proteins co-immunoprecipitating with the aPPR-bound psbA pointed to a set of ~22 proteins that were highly enriched in immunoprecipitates from both SCD11 and SCD14 plants. Among these proteins were several previously reported to interact with psbA. There were also proteins of unknown function, and the authors further analyzed two of these, SRRP1 and CP33C, both of which have predicted RNA binding domains.
RIP-seq experiments in maize (Zea mays) showed that the maize homologs of SRRP1 and CP33C indeed bind psbA in vivo, and do so quite specifically. Thus, this method of using a designer aPPR-affinity tag to pull down a specific RNA can successfully identify candidate interacting partners of that target RNA. The ability to identify proteins bound to an RNA represents an important advance for the field, in which much has been learned from characterizing RNAs bound to a specific protein.
REFERENCES
Barkan A, Rojas M, Fujii S, Yap A, Chong YS, Bond CS and Small I. (2012) A Combinatorial Amino Acid Code for RNA Recognition by Pentatricopeptide Repeat Proteins. PLOS Genetics, 8: e1002910.
Hall, T.M. (2016). De-coding and re-coding RNA recognition by PUF and PPR repeat proteins. Curr Opin Struct Biol 36: 116-121.
Helder, S., Blythe, A.J., Bond, C.S., and Mackay, J.P. (2016). Determinants of affinity and specificity in RNA-binding proteins. Curr Opin Struct Biol 38: 83-91.
McDermott, J.J., Watkins, K.P., Williams-Carrier, R., and Barkan, A. (2019). Ribonucleoprotein Capture by in vivo Expression of a Designer Pentatricopeptide Repeat Protein in Arabidopsis. Plant Cell. doi: 10.1105/tpc.19.00177. {https://doi.org/10.1105/tpc.19.00177}
Miranda, R.G., McDermott, J.J., and Barkan, A. (2018). RNA-binding specificity landscapes of designer pentatricopeptide repeat proteins elucidate principles of PPR-RNA interactions. Nucleic Acids Res 46: 2613-2623.




