Correcting organellar spelling with RNA editors
Oldenkott et al. use evolution to their advantage by testing whether independently evolved RNA editors can reciprocally correct the errors of organellar transcripts. The Plant Cell (2020) https://doi.org/10.1105/tpc.20.00311.
By Bastian Oldenkott and Mareike Schallenberg-Rüdinger, IZMB, University of Bonn, Germany
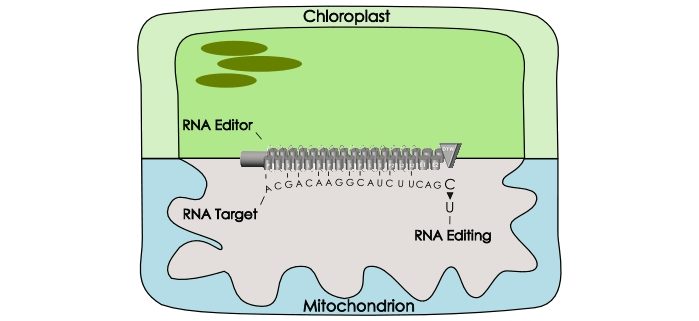
Background: Plant cells contain three genomes. Aside from the nuclear genome, mitochondria and chloroplasts also contain their own genomic DNA. These organelles are obviously critical for plant life, since they are the main cellular power stations. Surprisingly though, mitochondrial and plastid genes contain errors that must be corrected before producing functional proteins during translation. These errors are corrected at the transcript level by so-called RNA editors that are encoded by the nuclear genome, namely DYW-type Pentatricopeptide Repeat (PPR) proteins, which catalyze the exchange of cytidine bases (C) to uridine (U) (two of the four RNA building blocks). This process is called C-to-U RNA editing. Since the RNA editors are sequence-specific, various plant species have expanded their PPR protein family up to thousands of members. The moss Physcomitrium patens (formerly Physcomitrella patens) is the exact opposite: it is a plant with low RNA editing, with only 9 RNA editors and only 13 errors to correct, making it a simple model plant of choice.
Question: The mechanism of C-to-U RNA editing can be used to modify specific positions in transcribed RNAs. We were highly interested to test if individual RNA editors were interchangeable between plant species.
Findings: For our study, we chose species separated by over 400 million years of evolution: The moss P. patens and the flowering plants Arabidopsis thaliana and Macadamia integrifolia. These plants have one thing in common: They share the same error in their mitochondrial genome, corrected by species-specific editors.
When we transferred the RNA editors between these species, the P. patens RNA editor corrected the error in the flowering plant A. thaliana, but not the other way around. The M. integrifolia PPR protein, however, is functional in the P. patens and A. thaliana systems. These results demonstrate that at least some RNA editors work independently of their origin.
Next steps: We have shown that PPR-type RNA editors function across distant plant systems. As a next step, we wish to harness these RNA editors to introduce specific mutations in organellar genes of various plant species. This method provides a way to inactivate organellar genes and study their function, or to make chloroplasts and mitochondria work better.
Bastian Oldenkott et al. (2020). One C-to-U RNA editing site and two independently evolved editing factors: testing reciprocal complementation with DYW-type PPR proteins from the moss Physcomitrium (Physcomitrella) patens and the flowering plants Macadamia integrifolia and Arabidopsis thaliana. Plant Cell. DOI: https://doi.org/10.1105/tpc.20.00311