Complete microviscosity maps of living plant cells and tissues with a toolbox of targeting mechanoprobes (PNAS)
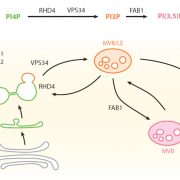
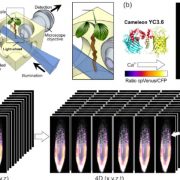
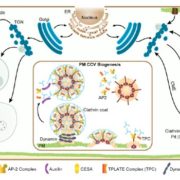
 Biological processes are ruled by physical as well as chemical forces and properties, but the former are much less well understood and studied. Here, Michels et al. describe a collection of fluorescent probes that provide a quantitative visual readout of microviscocity, reflecting the physical properties of cellular compartments. The core of each probe is a molecular rotor. Photoexcition causes rotation that leads to fluorescence, and the rate of decay of the excited state is determined by the time it takes for the probe to relax to the initial state – in other words, more viscosity or other rotational constraints leads to a longer emission period. The authors developed a set of probes with chemical modifications that target each to the vacuole, cytosol, membrane or cell wall. The decay time is affected differently by the different compositions of each compartment (for example, in membranes it is affected by lipid packing, and in walls by the network mesh size), but within a compartment the probes can detect localized differences in microviscosity. Using these probes, the authors demonstrate differences between cell types and during cell differentiation. As the authors summarize, “These results pave the way to the noninvasive micromechanical mapping of complex tissues.” (Summary by Mary Williams @PlantTeaching) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1921374117
Biological processes are ruled by physical as well as chemical forces and properties, but the former are much less well understood and studied. Here, Michels et al. describe a collection of fluorescent probes that provide a quantitative visual readout of microviscocity, reflecting the physical properties of cellular compartments. The core of each probe is a molecular rotor. Photoexcition causes rotation that leads to fluorescence, and the rate of decay of the excited state is determined by the time it takes for the probe to relax to the initial state – in other words, more viscosity or other rotational constraints leads to a longer emission period. The authors developed a set of probes with chemical modifications that target each to the vacuole, cytosol, membrane or cell wall. The decay time is affected differently by the different compositions of each compartment (for example, in membranes it is affected by lipid packing, and in walls by the network mesh size), but within a compartment the probes can detect localized differences in microviscosity. Using these probes, the authors demonstrate differences between cell types and during cell differentiation. As the authors summarize, “These results pave the way to the noninvasive micromechanical mapping of complex tissues.” (Summary by Mary Williams @PlantTeaching) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1921374117