Comparative transcriptomics in ferns provide a framework for their unique evolutionary path
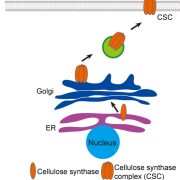
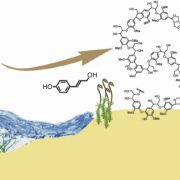
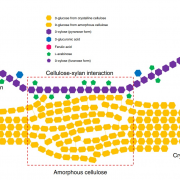
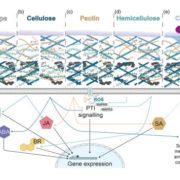
 Ferns are important and diverse land plants but are also known for their exceptionally large genomes. A new study by Ali et al. presents an extensive analysis of fern genomics through RNA-sequencing of 22 representative fern species. The study identified 18 whole-genome duplications across different fern lineages, contributing to their high chromosome numbers and species diversity. The analysis of fern gene functions shows that over 50% of gene families are fern-specific, indicating novel, unexplored functions that differentiate them from other land plants. The study also highlights the absence of several genes crucial for hormonal signaling, defense, and development found in angiosperms, reflecting ferns’ unique strategies for growth and survival. Additionally, the research revealed the presence of an unusual sugar, 2-O-methyl-D-glucopyranose, in certain fern species, suggesting a divergent evolutionary path in cell wall biochemistry. An investigation into fern cell walls identifies the presence of mixed-linkage glucans and variations in polysaccharide composition, underscoring the complexity and diversity of fern cell wall structures. (Summary by Amarachi Ezeoke) bioRxiv https://doi.org/10.1101/2024.08.27.609851
Ferns are important and diverse land plants but are also known for their exceptionally large genomes. A new study by Ali et al. presents an extensive analysis of fern genomics through RNA-sequencing of 22 representative fern species. The study identified 18 whole-genome duplications across different fern lineages, contributing to their high chromosome numbers and species diversity. The analysis of fern gene functions shows that over 50% of gene families are fern-specific, indicating novel, unexplored functions that differentiate them from other land plants. The study also highlights the absence of several genes crucial for hormonal signaling, defense, and development found in angiosperms, reflecting ferns’ unique strategies for growth and survival. Additionally, the research revealed the presence of an unusual sugar, 2-O-methyl-D-glucopyranose, in certain fern species, suggesting a divergent evolutionary path in cell wall biochemistry. An investigation into fern cell walls identifies the presence of mixed-linkage glucans and variations in polysaccharide composition, underscoring the complexity and diversity of fern cell wall structures. (Summary by Amarachi Ezeoke) bioRxiv https://doi.org/10.1101/2024.08.27.609851