Chloroplast Damage Triggers Autophagy
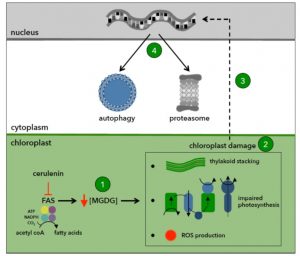 Under normal growth conditions, there is a constitutive or basal level of autophagy in plant cells that helps to recycle unnecessary cell contents. Autophagy provides essential building blocks (e.g. amino acids and fatty acids) and energy sources that promote cell homeostasis and survival. Autophagy is mediated by autophagy-related (ATG) genes, which have been identified in most eukaryotes including algae and plants. A subset of ATG proteins constitutes the autophagy core machinery and is essential for the formation of the autophagosome and its fusion to the vacuole. Unlike in plants, ATG genes are single copy in the Chlamydomonas genome, which facilitates the study of autophagy in this alga. Heredia-Martínez et al. (10.1104/pp.18.00630) have investigated the relationship between lipid metabolism and autophagy in Chlamydomonas by inhibiting the synthesis of fatty acids in the chloroplast with cerulenin, a specific inhibitor of type II fatty acid synthase (FAS). They report that FAS inhibition activates autophagy in Chlamydomonas. FAS inhibition decreased the content of monogalactosyldiacylglycerol (MGDG), the most abundant plastid lipid, and altered the ultrastructure and function of the chloroplast, leading to photo-oxidative damage and the activation of major catabolic pathways in the cell. The inhibition of fatty acid synthesis also increased the lutein content, downregulated photosynthesis, and increased the production of reactive oxygen species. Electron microscopy revealed a high degree of thylakoid membrane stacking in cerulenin-treated cells. Moreover, global transcriptomic analysis of these cells showed an upregulation of genes encoding chloroplast proteins involved in protein folding and oxidative stress. These results suggest a link between lipid metabolism, chloroplast integrity, and autophagy.
Under normal growth conditions, there is a constitutive or basal level of autophagy in plant cells that helps to recycle unnecessary cell contents. Autophagy provides essential building blocks (e.g. amino acids and fatty acids) and energy sources that promote cell homeostasis and survival. Autophagy is mediated by autophagy-related (ATG) genes, which have been identified in most eukaryotes including algae and plants. A subset of ATG proteins constitutes the autophagy core machinery and is essential for the formation of the autophagosome and its fusion to the vacuole. Unlike in plants, ATG genes are single copy in the Chlamydomonas genome, which facilitates the study of autophagy in this alga. Heredia-Martínez et al. (10.1104/pp.18.00630) have investigated the relationship between lipid metabolism and autophagy in Chlamydomonas by inhibiting the synthesis of fatty acids in the chloroplast with cerulenin, a specific inhibitor of type II fatty acid synthase (FAS). They report that FAS inhibition activates autophagy in Chlamydomonas. FAS inhibition decreased the content of monogalactosyldiacylglycerol (MGDG), the most abundant plastid lipid, and altered the ultrastructure and function of the chloroplast, leading to photo-oxidative damage and the activation of major catabolic pathways in the cell. The inhibition of fatty acid synthesis also increased the lutein content, downregulated photosynthesis, and increased the production of reactive oxygen species. Electron microscopy revealed a high degree of thylakoid membrane stacking in cerulenin-treated cells. Moreover, global transcriptomic analysis of these cells showed an upregulation of genes encoding chloroplast proteins involved in protein folding and oxidative stress. These results suggest a link between lipid metabolism, chloroplast integrity, and autophagy.



