What We’re Reading: October 20th
Review: DNA sequencing at 40: past, present and future ($)
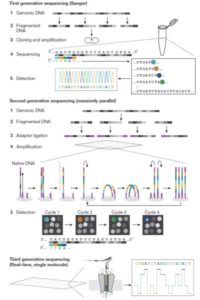 Shendure et al. provide a superb review of how DNA sequencing technology has changed over the years and how these changes open up new applications. They start with the Maxam and Gilbert chemical cleavage and the Sanger “chain-termination” methods developed in the 1970s, and describe the scale-ups required for the draft completion in 2001 of the Human Genome Project. These “first-generation” approaches were followed by the development of massively parallel or “next-generation” and “sequence-by-synthesis” methods in the 2000s, now dominated by the Illumina platform. We’re now in the era of “real-time, single-molecule” sequencing, exemplified by PacBio and Nanopore technologies. Although these single-molecule long reads are still prone to errors, they provide the essential longer scaffolds to which short reads can be aligned. This excellent review includes a timeline of genome milestones [including our favs, Arabidopsis thaliana (2000), Oryza sativa (2005), Cyanidioschyzon merolae (a red alga, 2007) and Zea mays (2009)] as well as computational and application milestones. The review ends wioth a look at the future technologies and applications of DNA sequencing, which the authors predict “will have a longevity and impact on par with or exceeding that of the microscope.” A super review article for teaching! Nature 10.1038/nature24286
Shendure et al. provide a superb review of how DNA sequencing technology has changed over the years and how these changes open up new applications. They start with the Maxam and Gilbert chemical cleavage and the Sanger “chain-termination” methods developed in the 1970s, and describe the scale-ups required for the draft completion in 2001 of the Human Genome Project. These “first-generation” approaches were followed by the development of massively parallel or “next-generation” and “sequence-by-synthesis” methods in the 2000s, now dominated by the Illumina platform. We’re now in the era of “real-time, single-molecule” sequencing, exemplified by PacBio and Nanopore technologies. Although these single-molecule long reads are still prone to errors, they provide the essential longer scaffolds to which short reads can be aligned. This excellent review includes a timeline of genome milestones [including our favs, Arabidopsis thaliana (2000), Oryza sativa (2005), Cyanidioschyzon merolae (a red alga, 2007) and Zea mays (2009)] as well as computational and application milestones. The review ends wioth a look at the future technologies and applications of DNA sequencing, which the authors predict “will have a longevity and impact on par with or exceeding that of the microscope.” A super review article for teaching! Nature 10.1038/nature24286
De novo assembly of a new Solanum pennellii accession using nanopore sequencing
 Chromosomes are long, and DNA sequencing reads have typically been short, meaning that it is necessary to assemble lots and lots of short reads by looking for overlapping sequences. This strategy is made more difficult in repeat-rich and transposon-rich regions of genomes, which characterize many plant genomes. The development of single-molecule sequencing like PacBio and Nanopore technologies provides a partial solution to that problem, by providing a backbone onto which these short reads can be mapped. Schmidt et al. took advantage of nanopore sequencing to sequence a new, resiliant accession of the wild tomato Solanum pennellii. The authors summarize, “Taken together our data indicate that such long read sequencing data can be used to affordably sequence and assemble gigabase-sized plant genomes.” Plant Cell 10.1105/tpc.17.00521
Chromosomes are long, and DNA sequencing reads have typically been short, meaning that it is necessary to assemble lots and lots of short reads by looking for overlapping sequences. This strategy is made more difficult in repeat-rich and transposon-rich regions of genomes, which characterize many plant genomes. The development of single-molecule sequencing like PacBio and Nanopore technologies provides a partial solution to that problem, by providing a backbone onto which these short reads can be mapped. Schmidt et al. took advantage of nanopore sequencing to sequence a new, resiliant accession of the wild tomato Solanum pennellii. The authors summarize, “Taken together our data indicate that such long read sequencing data can be used to affordably sequence and assemble gigabase-sized plant genomes.” Plant Cell 10.1105/tpc.17.00521
High contiguity Arabidopsis thaliana genome assembly with a single nanopore flow cell
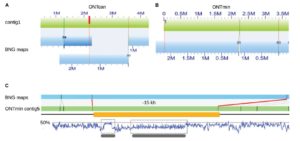 The current version of the Arabidopsis thaliana Col-0 reference genome, TAIR10, still has some gaps and mis-assemblies due to centromeres and repeat-rich regions. In another demonstration of the promise of single-molecule sequencing, Michael, Jupe et al. used a single nanopore flow cell to sequence the Arabidopsis thaliana accession KBS-Mac-74 in “four days at a cost of less than 1,000 USD for sequencing consumables including instrument depreciation.” bioRxiv 10.1101/149997
The current version of the Arabidopsis thaliana Col-0 reference genome, TAIR10, still has some gaps and mis-assemblies due to centromeres and repeat-rich regions. In another demonstration of the promise of single-molecule sequencing, Michael, Jupe et al. used a single nanopore flow cell to sequence the Arabidopsis thaliana accession KBS-Mac-74 in “four days at a cost of less than 1,000 USD for sequencing consumables including instrument depreciation.” bioRxiv 10.1101/149997
Genome of wild olive and the evolution of oil biosynthesis
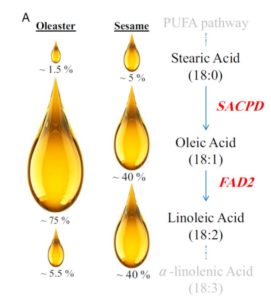 Olive oil is a staple of the healthy “Mediterranean diet” and contains high levels of the monounsaturated fat oleic acid. Unver, Wu et al. present the genome of the wild olive tree (Olea europaea var. sylvestris) (draft sequences of domesticated olive trees without extensive functional annotation were reported previously). This new work also looks at gene expression levels and investigates oil biosynthetic pathway genes. The authors find that changes in expression of genes encoding key enzymes in fatty acid synthesis (SACPD and FAD2) likely underlie oleic acid abundance in olive oil. Proc. Natl. Acad. Sci. USA 10.1073/pnas.1708621114
Olive oil is a staple of the healthy “Mediterranean diet” and contains high levels of the monounsaturated fat oleic acid. Unver, Wu et al. present the genome of the wild olive tree (Olea europaea var. sylvestris) (draft sequences of domesticated olive trees without extensive functional annotation were reported previously). This new work also looks at gene expression levels and investigates oil biosynthetic pathway genes. The authors find that changes in expression of genes encoding key enzymes in fatty acid synthesis (SACPD and FAD2) likely underlie oleic acid abundance in olive oil. Proc. Natl. Acad. Sci. USA 10.1073/pnas.1708621114
The genome of Quenopodium quinoa, a halophytic pseudocereal
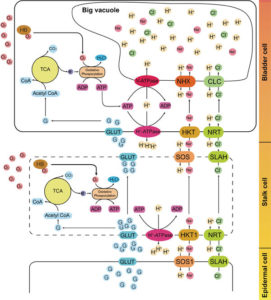 Quenopodiium quinoa is a highly nutritive and facultative halophyte pseudocereal whose cultivation has increased 10 fold in the last decades. However, the adaption to non-native areas is not easy to achieve and the limited genetic resources do not allow a breeding program. Zou and collaborators have generated a genome draft, covering 90.2% of the nuclear genome, with 54,438 protein-coding genes. Of these protein-coding genes, 95.3% were functionally annotated. Stress related genes, i.e., ABA signaling and ABA biosynthesis, had higher gene copy number compared to other species. The authors also analyzed the transcriptome of the epidermal bladder cells (EBCs, a specialized cell that sequesters salts from surrounding cells) and compared it to the rest of leaf cells. Genes involved in abiotic stress response, cell wall and suberin synthesis were upregulated in EBCs. In addition, using the results of the EBC transcriptome analysis, the authors propose a model for bladder salt accumulation. (Summary by Cecelia Vasquez-Robinet) Cell Research 10.1038/cr.2017.124
Quenopodiium quinoa is a highly nutritive and facultative halophyte pseudocereal whose cultivation has increased 10 fold in the last decades. However, the adaption to non-native areas is not easy to achieve and the limited genetic resources do not allow a breeding program. Zou and collaborators have generated a genome draft, covering 90.2% of the nuclear genome, with 54,438 protein-coding genes. Of these protein-coding genes, 95.3% were functionally annotated. Stress related genes, i.e., ABA signaling and ABA biosynthesis, had higher gene copy number compared to other species. The authors also analyzed the transcriptome of the epidermal bladder cells (EBCs, a specialized cell that sequesters salts from surrounding cells) and compared it to the rest of leaf cells. Genes involved in abiotic stress response, cell wall and suberin synthesis were upregulated in EBCs. In addition, using the results of the EBC transcriptome analysis, the authors propose a model for bladder salt accumulation. (Summary by Cecelia Vasquez-Robinet) Cell Research 10.1038/cr.2017.124
Improving crops by genome editing
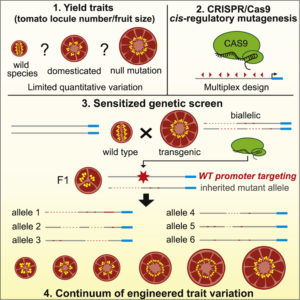 Introducing new desirable traits in domesticated crops takes time and is limited by the need for existing trait variation in members of the same species or closely related species. Rodríguez-Leal et al. propose a system by which variation in a quantitative trait can be generated by editing the promoter (where numerous transcriptional regulatory sequences can be found) of genes of interest. Using a clever CRISPR/Cas9 “toolkit”, the authors produced phenotypic variation in developmental genes that influence final yields, all without carrying any transgene in the final new crop variant. This method sets an exciting future for crop breeding. (Summary by Gaby Auge) Cell 10.1016/j.cell.2017.08.030
Introducing new desirable traits in domesticated crops takes time and is limited by the need for existing trait variation in members of the same species or closely related species. Rodríguez-Leal et al. propose a system by which variation in a quantitative trait can be generated by editing the promoter (where numerous transcriptional regulatory sequences can be found) of genes of interest. Using a clever CRISPR/Cas9 “toolkit”, the authors produced phenotypic variation in developmental genes that influence final yields, all without carrying any transgene in the final new crop variant. This method sets an exciting future for crop breeding. (Summary by Gaby Auge) Cell 10.1016/j.cell.2017.08.030
Expression and purification of unstable proteins using an ASK-assisted system
 Ubiquitin-mediated protein degradation is important in many plant processes (growth, development, responses to stress) but little is known about F-box proteins, a key component of the SCF (SKP1-CUL1-F-box protein) E3 ubiquitin ligase complex, due to their unstable protein structures. Li et al. investigated the use of ASK1 (Arabidopsis SKP1-like1, which binds to SCF E3 ubiquitin ligase complexes) to aid in the expression and purification of F-box proteins (COI1, TIR1, SLY1 and EBF1) in a baculovirus–insect cell expression system. They show that ASK1 improves the expression of F-box proteins, that ASK1 is essential for maintaining the bioactivity of the F-box proteins, and that different ASK proteins have different effects on F-box protein expression. The authors also describe a novel F-box protein purification process to generate high purity F-box proteins. (Summary by Julia Miller) Plant J. 10.1111/tpj.13708
Ubiquitin-mediated protein degradation is important in many plant processes (growth, development, responses to stress) but little is known about F-box proteins, a key component of the SCF (SKP1-CUL1-F-box protein) E3 ubiquitin ligase complex, due to their unstable protein structures. Li et al. investigated the use of ASK1 (Arabidopsis SKP1-like1, which binds to SCF E3 ubiquitin ligase complexes) to aid in the expression and purification of F-box proteins (COI1, TIR1, SLY1 and EBF1) in a baculovirus–insect cell expression system. They show that ASK1 improves the expression of F-box proteins, that ASK1 is essential for maintaining the bioactivity of the F-box proteins, and that different ASK proteins have different effects on F-box protein expression. The authors also describe a novel F-box protein purification process to generate high purity F-box proteins. (Summary by Julia Miller) Plant J. 10.1111/tpj.13708
The Cys-Arg/N-end rule pathway is a general sensor of abiotic stress in flowering plants
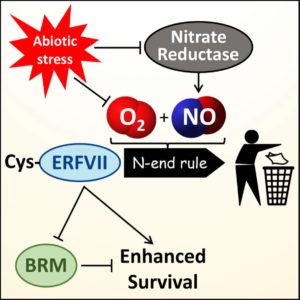 The N-end rule pathway is a conserved pathway for the control of protein turnover, through which the clipping or modification of amino acids from the amino-terminus of a protein leads to an interaction with PROTEOLYSIS 6 (PRT6; an N-recognin E3 ligase) and 26S proteasome-mediated proteolysis. Previously, hypoxia (low oxygen) and nitric oxide (NO) were shown to be sensed by the oxidation state of an N-terminal Cys residue of a subset of ERF transcription factors known as ERFVII. Vicente, Mendiondo et al. show other abiotic stresses are sensed through N-rule pathway regulation of ERFVII levels. Barley and Arabidopsis plants with constitutively-stabilized ERFVIIs (e.g., through loss of activity of PRT6) show enhanced survival when challenged with heat, salt, drought and oxidative stresses, whereas mutant depleted in ERFVII activity showed less stress tolerance. These results indicate that ERFVIIs are stabilized during stress, and the authors showed that NO levels are decreased during stress due to a decrease in nitrate reductase activity. These and other findings suggest that, “stabilized ERFVIIs contribute to multiple protein-network hubs that balance growth, development, and response to environmental stresses.” Curr. Biol. 10.1016/j.cub.2017.09.006
The N-end rule pathway is a conserved pathway for the control of protein turnover, through which the clipping or modification of amino acids from the amino-terminus of a protein leads to an interaction with PROTEOLYSIS 6 (PRT6; an N-recognin E3 ligase) and 26S proteasome-mediated proteolysis. Previously, hypoxia (low oxygen) and nitric oxide (NO) were shown to be sensed by the oxidation state of an N-terminal Cys residue of a subset of ERF transcription factors known as ERFVII. Vicente, Mendiondo et al. show other abiotic stresses are sensed through N-rule pathway regulation of ERFVII levels. Barley and Arabidopsis plants with constitutively-stabilized ERFVIIs (e.g., through loss of activity of PRT6) show enhanced survival when challenged with heat, salt, drought and oxidative stresses, whereas mutant depleted in ERFVII activity showed less stress tolerance. These results indicate that ERFVIIs are stabilized during stress, and the authors showed that NO levels are decreased during stress due to a decrease in nitrate reductase activity. These and other findings suggest that, “stabilized ERFVIIs contribute to multiple protein-network hubs that balance growth, development, and response to environmental stresses.” Curr. Biol. 10.1016/j.cub.2017.09.006
Regulation of rice root development by a retrotransposon acting as a microRNA sponge
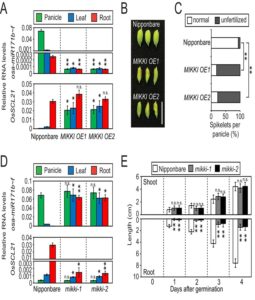 Transposable elements are known to affect their neighboring genes, but they were so far unknown to affect the function of distant genes. Cho and Paszkowski observed a high level of transposon transcription expressed in rice. A transposon with root specific expression, MIKKI, was found to contain an imperfect binding site for miRNA171, which in turn is known to target mRNAs of SCARECROW-like (SCL) transcription factors. Transgenic lines overexpressing MIKKI showed higher levels of SCL expression, resulting in abnormal flower development and sterility. On the other hand, lines where the recognition site for miRNA171 in MIKKI was mutated showed reduced SCL expression and reduced root length. This is the first study to show that TE-derived RNA acts as a molecular sponge for miRNA and affects plant development. (Summary by Magdalena Julkowska) eLIFE 10.7554/eLife.30038
Transposable elements are known to affect their neighboring genes, but they were so far unknown to affect the function of distant genes. Cho and Paszkowski observed a high level of transposon transcription expressed in rice. A transposon with root specific expression, MIKKI, was found to contain an imperfect binding site for miRNA171, which in turn is known to target mRNAs of SCARECROW-like (SCL) transcription factors. Transgenic lines overexpressing MIKKI showed higher levels of SCL expression, resulting in abnormal flower development and sterility. On the other hand, lines where the recognition site for miRNA171 in MIKKI was mutated showed reduced SCL expression and reduced root length. This is the first study to show that TE-derived RNA acts as a molecular sponge for miRNA and affects plant development. (Summary by Magdalena Julkowska) eLIFE 10.7554/eLife.30038
Small peptides, big importance: Small, secreted peptides as novel regulators of symbiosis and nutrient acquisition
 It is becoming increasingly evident that small, secreted peptides (SSPs) are important regulators of plant development and responses to stress. Traditional gene prediction algorithms are biased toward larger coding sequences and have therefore been inadequate in the hunt for plant SSPs. To address this gap in knowledge, de Bang et al. developed improved SSP prediction tools and reanalyzed 144 Medicago truncatula transcriptomes representing various abiotic and biotic stress conditions. SSPs with predicted importance in nutrient acquisition and bacterial symbiosis were validated by applying synthetic peptides to plants roots. This work reaffirms the importance of SSPs in plants and provides an improved computational framework for the prediction of functionally-relevant peptides. (Summary by Philip Carella) Plant Physiol. 10.1104/pp.17.01096
It is becoming increasingly evident that small, secreted peptides (SSPs) are important regulators of plant development and responses to stress. Traditional gene prediction algorithms are biased toward larger coding sequences and have therefore been inadequate in the hunt for plant SSPs. To address this gap in knowledge, de Bang et al. developed improved SSP prediction tools and reanalyzed 144 Medicago truncatula transcriptomes representing various abiotic and biotic stress conditions. SSPs with predicted importance in nutrient acquisition and bacterial symbiosis were validated by applying synthetic peptides to plants roots. This work reaffirms the importance of SSPs in plants and provides an improved computational framework for the prediction of functionally-relevant peptides. (Summary by Philip Carella) Plant Physiol. 10.1104/pp.17.01096
Highly expressed genes are preferentially co-opted for C4 photosynthesis
 One of the great questions of biology is how and why C4 photosynthesis pathway evolved independently more than 60 times. The advantages are obvious (increased productivity), but the underlying molecular predisposition to this transition remains poorly defined. Using a comparative transcriptomics approach involving the non-C4 relatives of C4 plants, Moreno-Villena et al. found that the members of gene families that were co-opted for C4 photosynthesis were those that were more highly expressed in the non-C4 ancestors. “The co-option, likely dictated by the mere presence of each protein in leaves, was followed by massive upregulation and widespread adaptation of coding sequences.” Mol. Biol. Evol. 10.1093/molbev/msx269
One of the great questions of biology is how and why C4 photosynthesis pathway evolved independently more than 60 times. The advantages are obvious (increased productivity), but the underlying molecular predisposition to this transition remains poorly defined. Using a comparative transcriptomics approach involving the non-C4 relatives of C4 plants, Moreno-Villena et al. found that the members of gene families that were co-opted for C4 photosynthesis were those that were more highly expressed in the non-C4 ancestors. “The co-option, likely dictated by the mere presence of each protein in leaves, was followed by massive upregulation and widespread adaptation of coding sequences.” Mol. Biol. Evol. 10.1093/molbev/msx269
Light scattering by floral nanostructures enhances signalling to bees
 Visual and other cues attract pollinators. Bees vision is skewed towards blue colors, but they also visit non-blue flowers. Moyroud et al. looked at how petal surface textures affect bee responses. The authors observed that similar parallel cuticular striations in diverse angiosperm lineages show convergent optical properties, and cause preferential scattering of short-wavelength blue and UV light. They mimicked this effect in artificial flowers and showed that the blue light attracts bees. Interestingly, perfectly ordered striations are less effective at blue-light scattering, and the conserved degree of disorder present in floral nanostructures increases their scattering intensity. Nature 10.1038/nature24285
Visual and other cues attract pollinators. Bees vision is skewed towards blue colors, but they also visit non-blue flowers. Moyroud et al. looked at how petal surface textures affect bee responses. The authors observed that similar parallel cuticular striations in diverse angiosperm lineages show convergent optical properties, and cause preferential scattering of short-wavelength blue and UV light. They mimicked this effect in artificial flowers and showed that the blue light attracts bees. Interestingly, perfectly ordered striations are less effective at blue-light scattering, and the conserved degree of disorder present in floral nanostructures increases their scattering intensity. Nature 10.1038/nature24285
Evidence for mid-Holocene rice domestication in the Americas
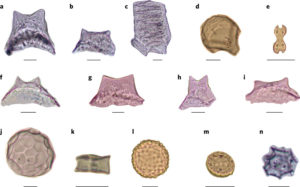 Rice is one of our most important crops, and previous work has indicated that it was domesticated independently in Asia (Oryza sativa) and Africa (Oryza glaberrima). Using archeological approaches, Hilbert et al. for the first time show evidence for domestication of a rice species (Oryza sp.) in the Americas. From excavation sites in Brazil, the authors obtained phytoliths (literally, “plant stones”), which are distinctive and characteristic silica deposits that form in plant tissues. Previous studies have shown that as plant sizes increase as a result of domestication, so do the sizes of phytoliths. The authors showed that not only did phytolith size increase but so did the proportion of rice phytoliths as compared to all phytoliths, and the proportion of husk to seed+stem phytoliths, indicating an increasing reliance on rice in the diet and better harvesting strategies. Nature Ecol. Evol. 10.1038/s41559-017-0322-4
Rice is one of our most important crops, and previous work has indicated that it was domesticated independently in Asia (Oryza sativa) and Africa (Oryza glaberrima). Using archeological approaches, Hilbert et al. for the first time show evidence for domestication of a rice species (Oryza sp.) in the Americas. From excavation sites in Brazil, the authors obtained phytoliths (literally, “plant stones”), which are distinctive and characteristic silica deposits that form in plant tissues. Previous studies have shown that as plant sizes increase as a result of domestication, so do the sizes of phytoliths. The authors showed that not only did phytolith size increase but so did the proportion of rice phytoliths as compared to all phytoliths, and the proportion of husk to seed+stem phytoliths, indicating an increasing reliance on rice in the diet and better harvesting strategies. Nature Ecol. Evol. 10.1038/s41559-017-0322-4